Happy to release Stacks v2.67 today. This release updates process_radtags to make it easier to use SRA data and to filter poly-G (error) runs coming from Nextseq/Novaseq machines + bugfixes. We also added a genotype depth filter to populations #RADseqcatchenlab.life.illinois.edu/stacks/
Thanks to Weixuan Ning, Heidi Meudt & @jentate01.bsky.social#AppsPlantSci#Polyploidybsapubs.onlinelibrary.wiley.com/doi/full/10....#plantjoy#botany#phylogenomics
Congruent patterns of cryptic cladogenesis revealed using RADseq and Sanger sequencing in a velvet worm species complex (Onychophora: Peripatopsidae: Peripatopsis sedgwicki) http://dlvr.it/T8l9D8
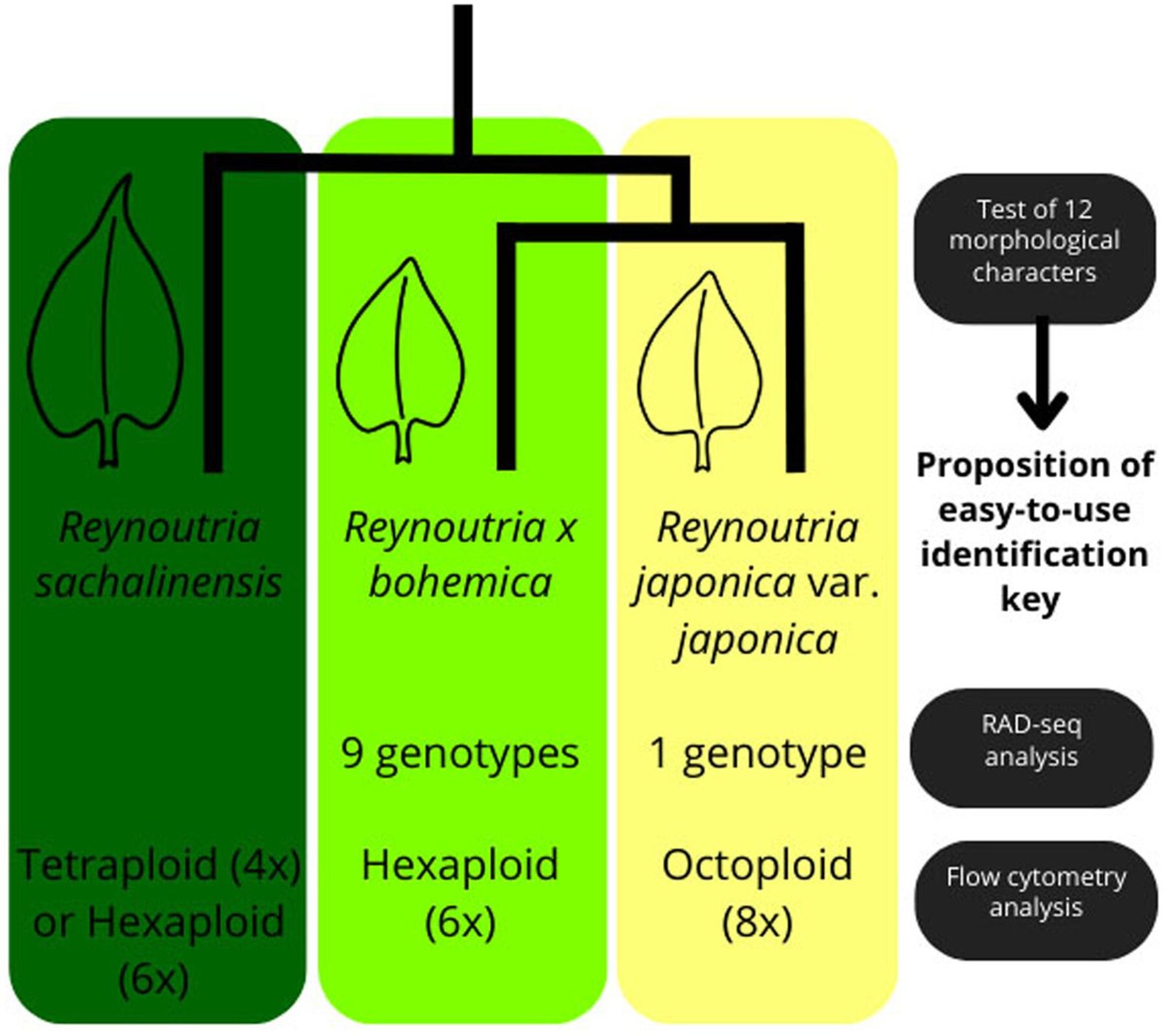
Untangling a knot of hybrids! Jugieau et al. use genetic, cytological, and morphological data to attempt to differentiate Asian knotweeds in northeastern France. #PlantScience#evolution#ecologyonlinelibrary.wiley.com/doi/10.1111/...
🚨Very happy to inform you that our #RADseq@naiarare.bsky.social Tereza and Natalia 🤩 ✏️https://physalia-courses.org/courses-workshops/course16/
My new paper with Liz Zimmer, @jlandisbotany.bsky.social@fernway.bsky.socialacademic.oup.com/aob/advance-...
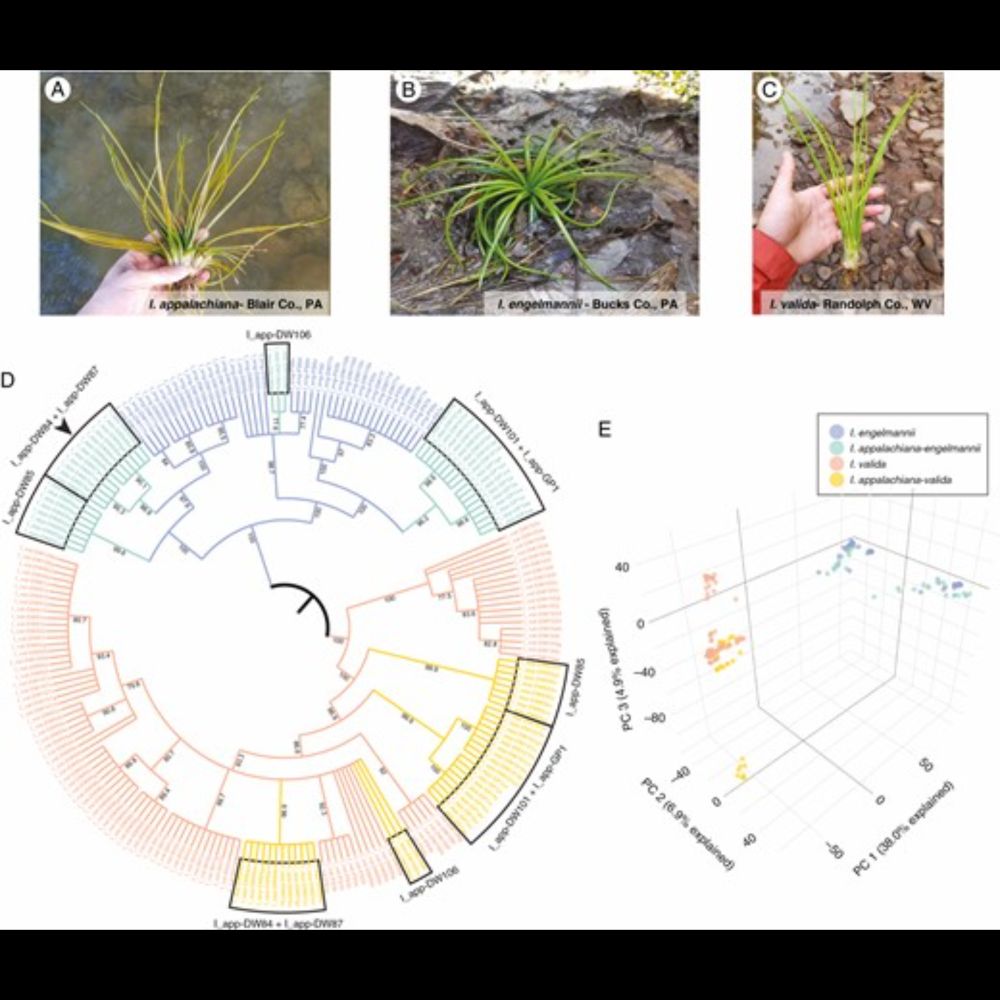
AbstractBackground and Aims. Allopolyploidy is an important driver of diversification and a key contributor to genetic novelty across the tree of life. However,
Kudos to this outstanding team of instructors @TerezaManousaki, @naiarare.bsky.social#RADseq data analysis. Their expertise has empowered our group of attendees, leaving them better equipped to navigate their own data.

It’s been a very intense week of coding around here in the RADseq Physalia course 🧠🔥 Can’t wait to analyze my own data 🧬