
is the most likely grouping? can be made a well-defined question. Note that the result that Cellstates returns is just the best partition it could find. In principle there is a whole posterior distribution over partitions. But in practice we have found that this posterior is very sharply peaked. n/n
This is a fundamental property of quantities whose measurements are noisy. Given two measured objects one cannot say that they either are the same or not, one can only calculate how (un)likely it is that they are the same. But the question: Assuming all objects in a group must be the same, what 1/n
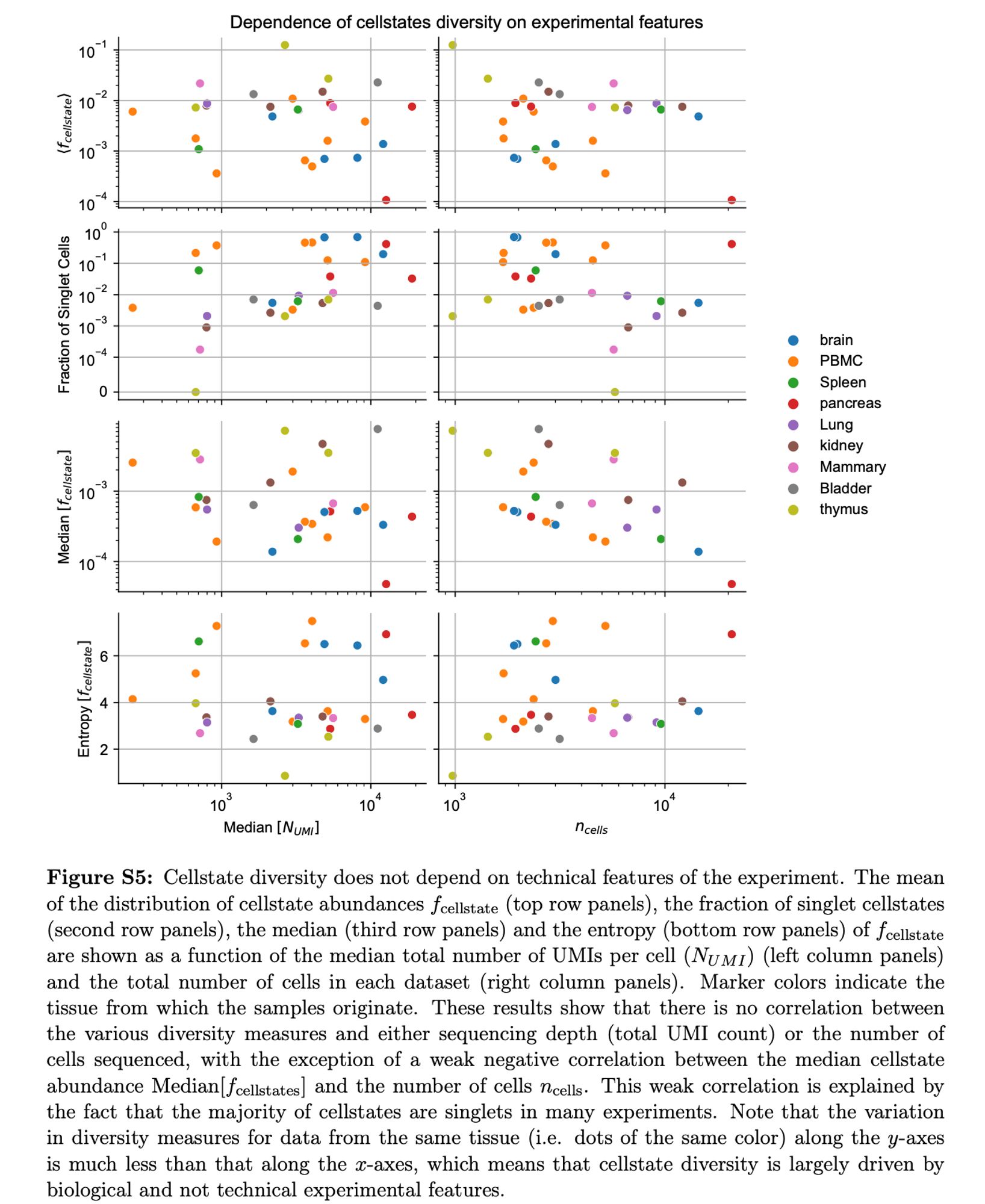
See Supplementary figure S5 of the paper. Singlet fraction does not correlate with either median UMI count or cell number (PBMCs are in orange). This was also to our surprise.
ie. other researchers will know that there was no opportunity to manipulate the results. We hope that this `slow analysis movement' will gain support in the field and that Cellstates will become one of the standard tools in its toolbox. n/n
and then carefully prepare a method that performs exactly that analysis without any extra bells, whistles, filters, or whatever other tunable parameters. This type of analysis may be slow, but it will have to be done only once. Moreover, its results will be unambiguous, 18/n
we would like to argue for a SLOW ANALYSIS movement. That is, instead of attempting many different kinds of analysis in rapid succession, until one gets results that one likes, we argue that one should carefully think before hand about exactly what analysis one wants to do, 17/n
We believe that this way of doing analysis is very unhealthy. Just like fast food is mostly unhealthy, and there is a `slow food' movement arguing for the benefits of food prepared and appreciated with more care and time devoted to it. Similarly, 16/n
field perform analysis in an almost trial-and-error manner, re-running many different tools many times, and tweaking all kinds of tunable parameters, cut-offs, and filters, until the results start aligning with preconceived ideas of what they expect to see in their data. 15/n
intend to address in a second version of the tool). We realize this high computational cost is going against the general trend in the field of wanting fast tools. However, we believe that this desire for fast analysis stems mainly from the fact that many researchers in the 14/n
Finally, I want to discuss one elephant in the room. Cellstates is computationally very expensive to run, especially for large datasets, and for very large datasets one would need to run Cellstates first on subsamples of the data to make it feasible (this is something we 13/n
