Just a clarification: I meant extracellular degradation of polysaccharides, not specifically of eps. Sorry about that.
Special thanks to all the co-autors: Christina Kaiser @christinakaiser.bsky.social@moritzmohrlok.bsky.social, Lauren Alteio, Shaul Polak and Hannes Schmidt !
We reveal how substrate concentration and complexity shape microbial growth and interactions. We also find data to support our model: we performed a systematic literature search, which shows that generalists (producing both enzymes) dominate among chitin-degrading microbes. 5/6
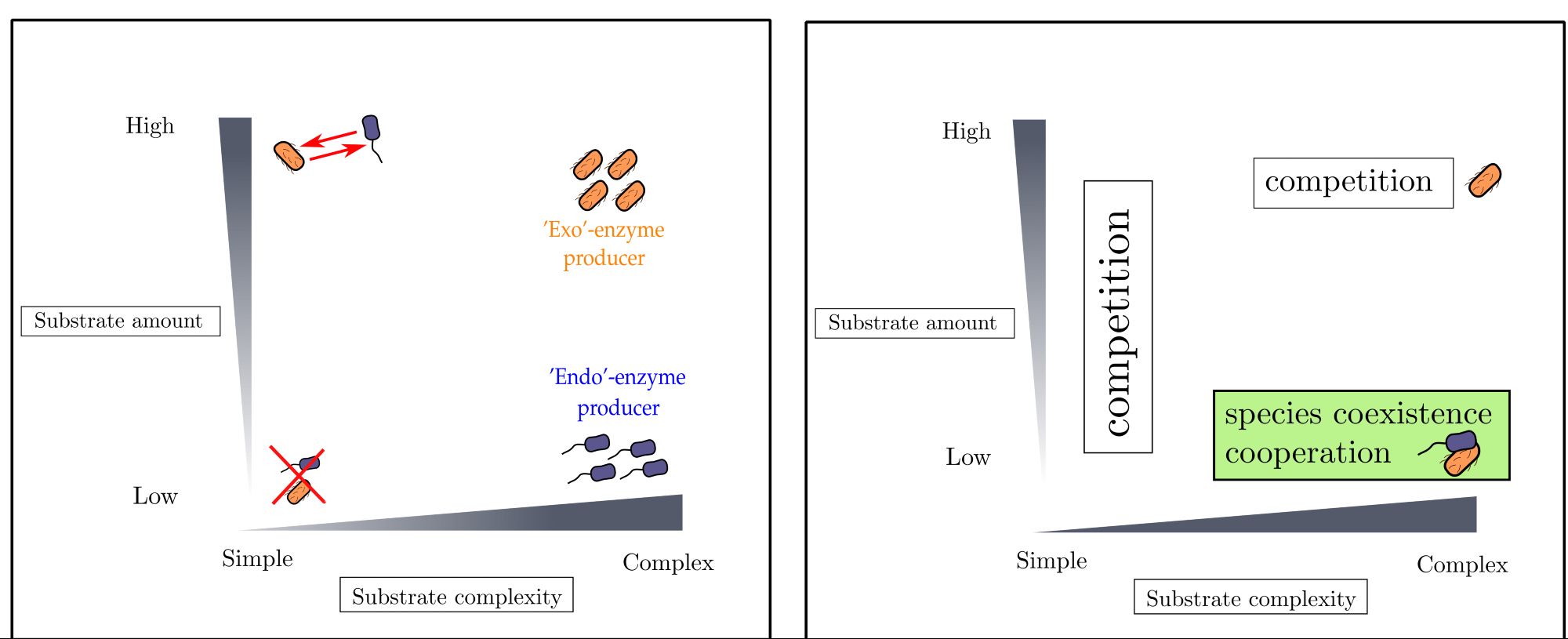
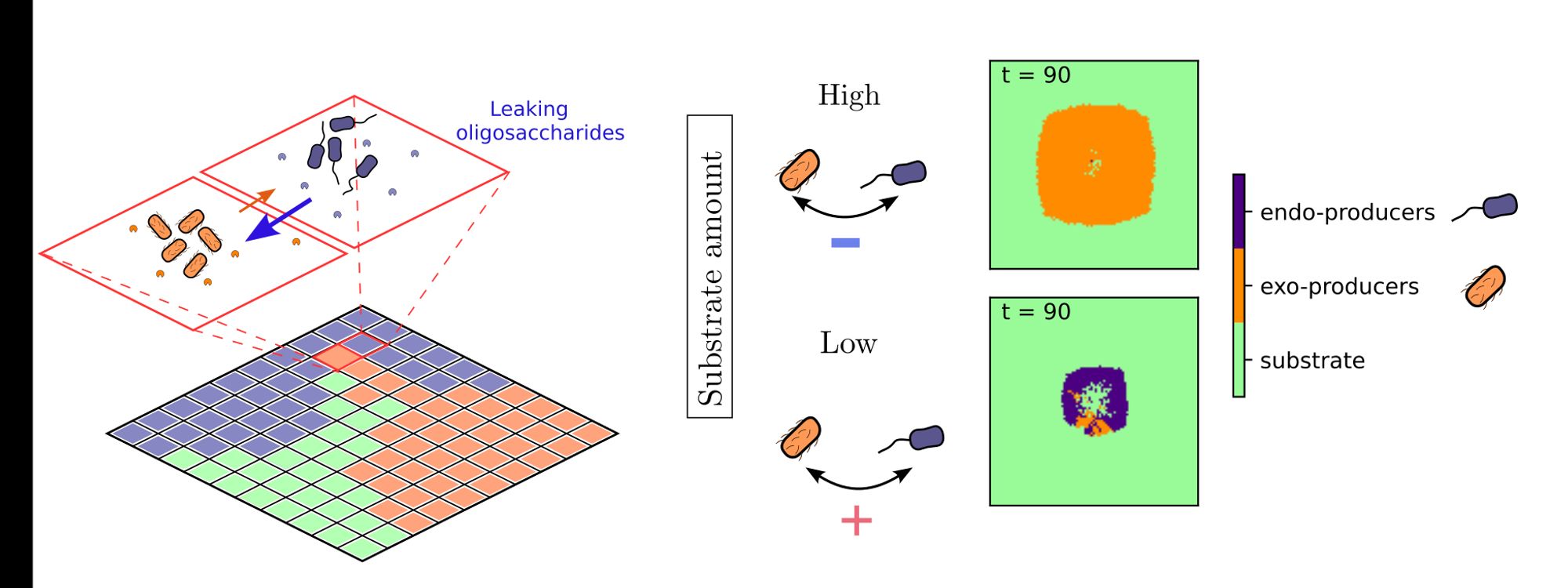
Finally, we explored whether microbes can divide labor in the production of these two enzymes within consortia. Our analysis paints a fascinating picture: interactions between endo- and exo-enzyme producers can flip from positive to negative depending on substrate levels! 4/6
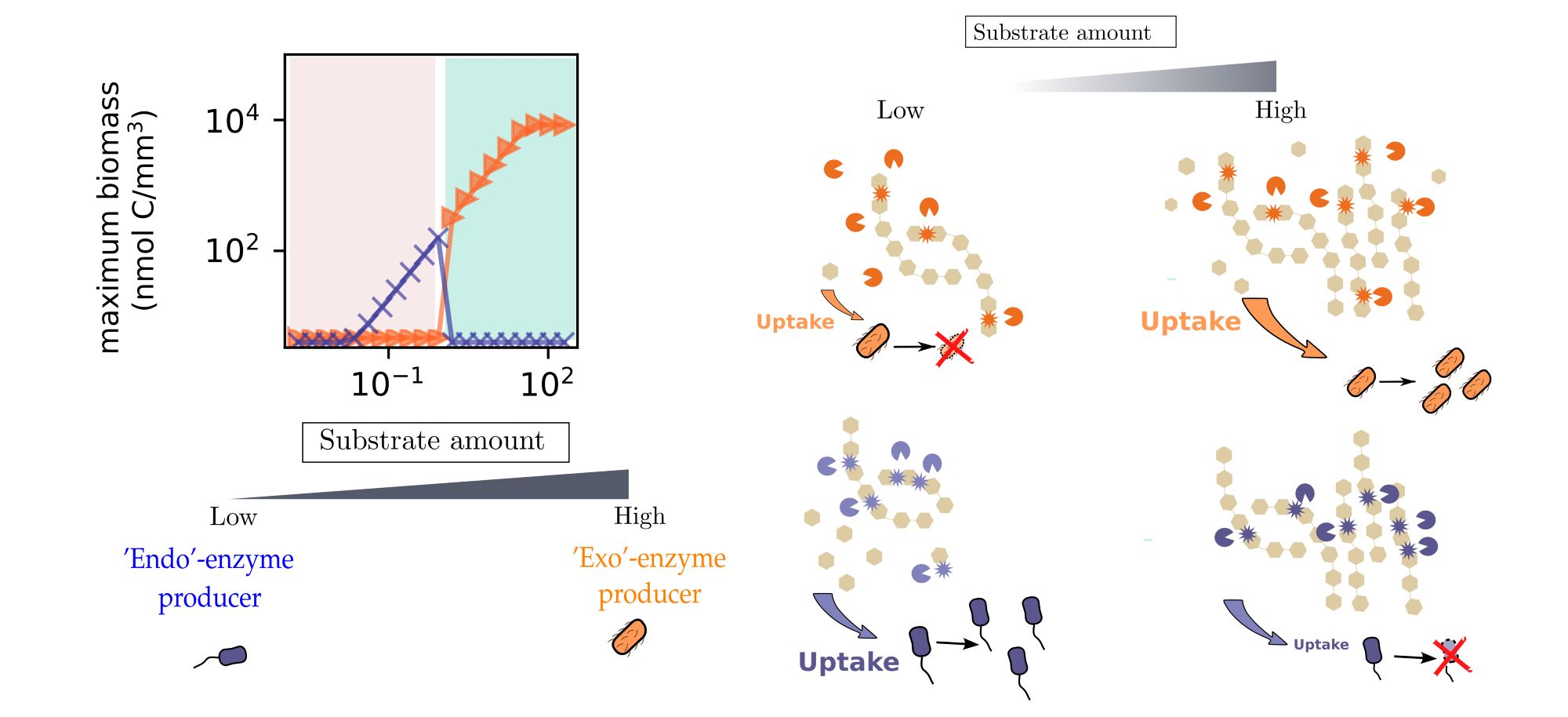
We have found that producers: 1. Of exo-enzymes thrive when many polymer chains are around. 2. Of endo-enzymes unexpectedly excel only when fewer chains are present. 3. Of both enzymes together expand their niche and may benefit from enzyme synergy. 3/6
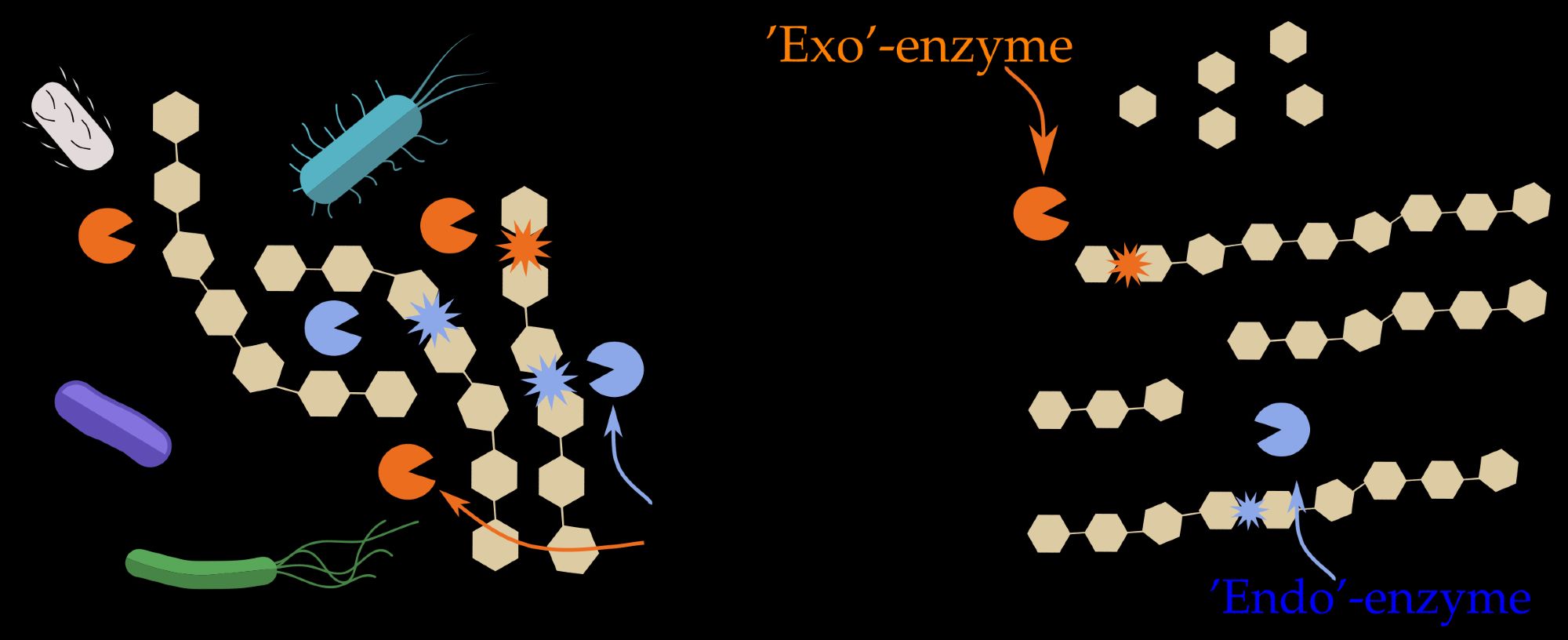
We mainly focus on two enzyme types: 1. “Exo”-enzymes snip small units from the ends of polymer chains. 2. “Endo”-enzymes break polymers at random, creating varied-sized fragments. Using a theoretical model, we simulated polymer degradation and microbial growth. 2/6
Excited to share our latest paper in PLOSCompBiol @plos.bsky.socialjournals.plos.org/ploscompbiol...#Microbialecology