Exciting work from Jinyoung Kim and Nick Ingolia enabling pooled CRISPRi screens in human cells by barcoded RNA readout. www.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
So, synonymous is not exactly the same, and the impact of codon choice continues to surprise us. Tomorrow is the fifth anniversary of starting my faculty job (and my birthday!) and despite pandemics and everything else, it's been a joy.
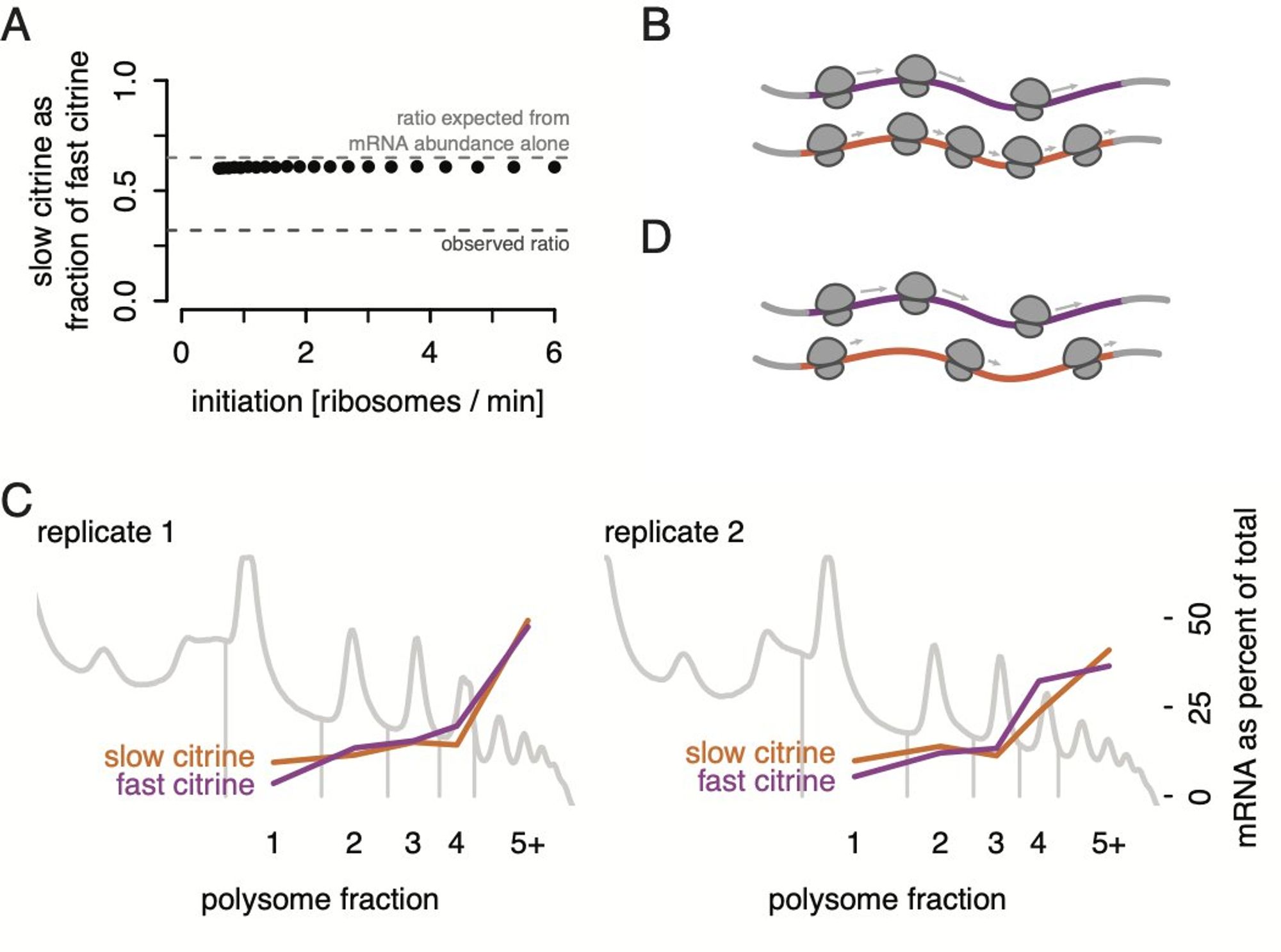
While we were digging into this, others have published some really cool results: low initiation in response to stalled ribosomes in mammalian cells (Weissman, Green, Kostova, Hegde labs) and strong evidence of less initiation on slow transcripts (Rissland lab). www.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
There must be some mechanism telling ribosomes not to start translating when things are moving too slowly. We haven’t pinned it down, but we’ve ruled out everything else. It was fun, and we’re excited to see if eIF3 is the next piece of the puzzle.
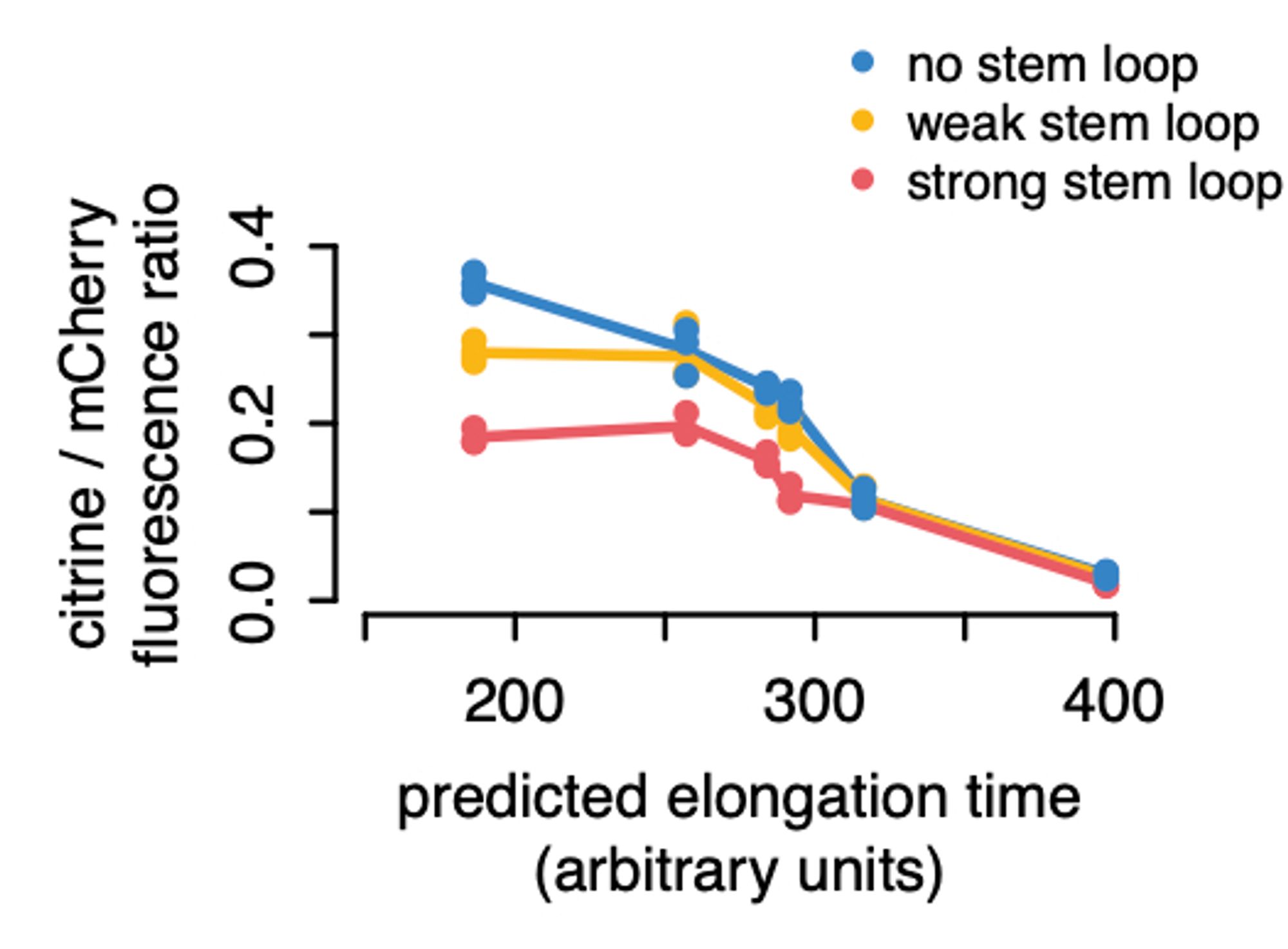
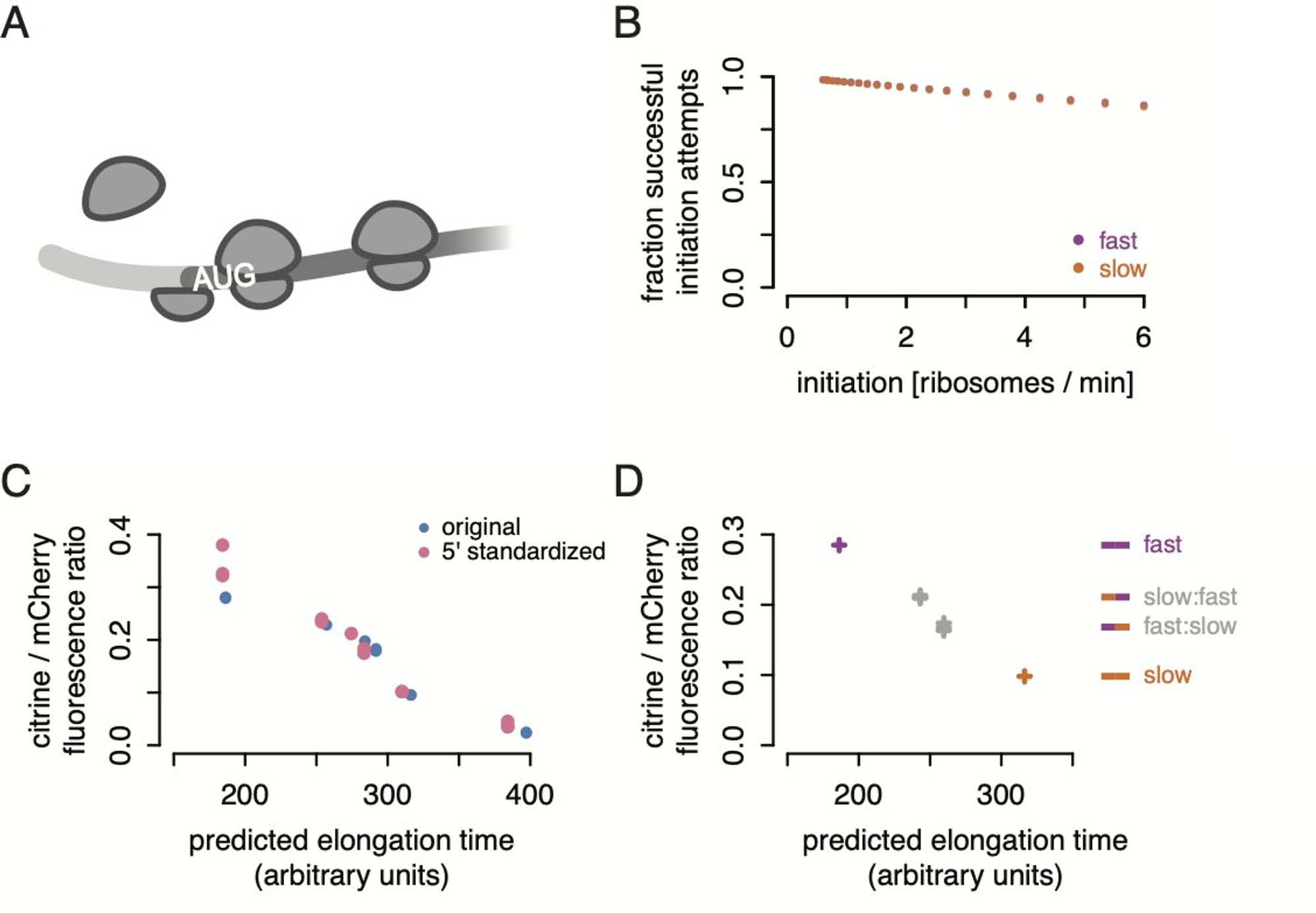
If you lower initiation, slow codons don’t matter as much, which we confirmed by sticking stem-loop structures in the 5’ UTRs. But we think there’s more to it than that.
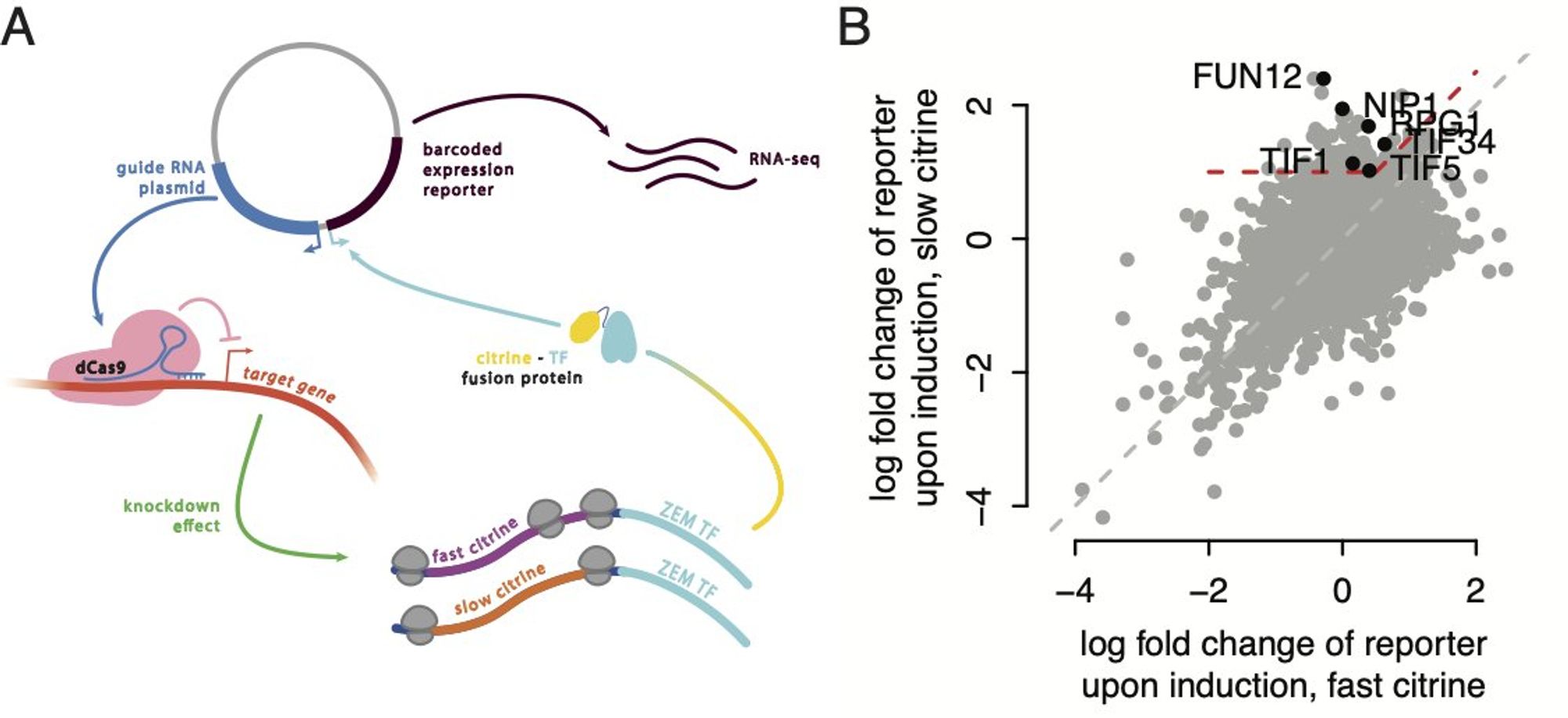
That pretty much left one option, some sort of active feedback on initiation. We did a CRISPRi screen using a version of the Ingolia lab’s CiBER-seq barcoded readout, and the one trend that came out was translation initiation factors, especially many subunits of eIF3.
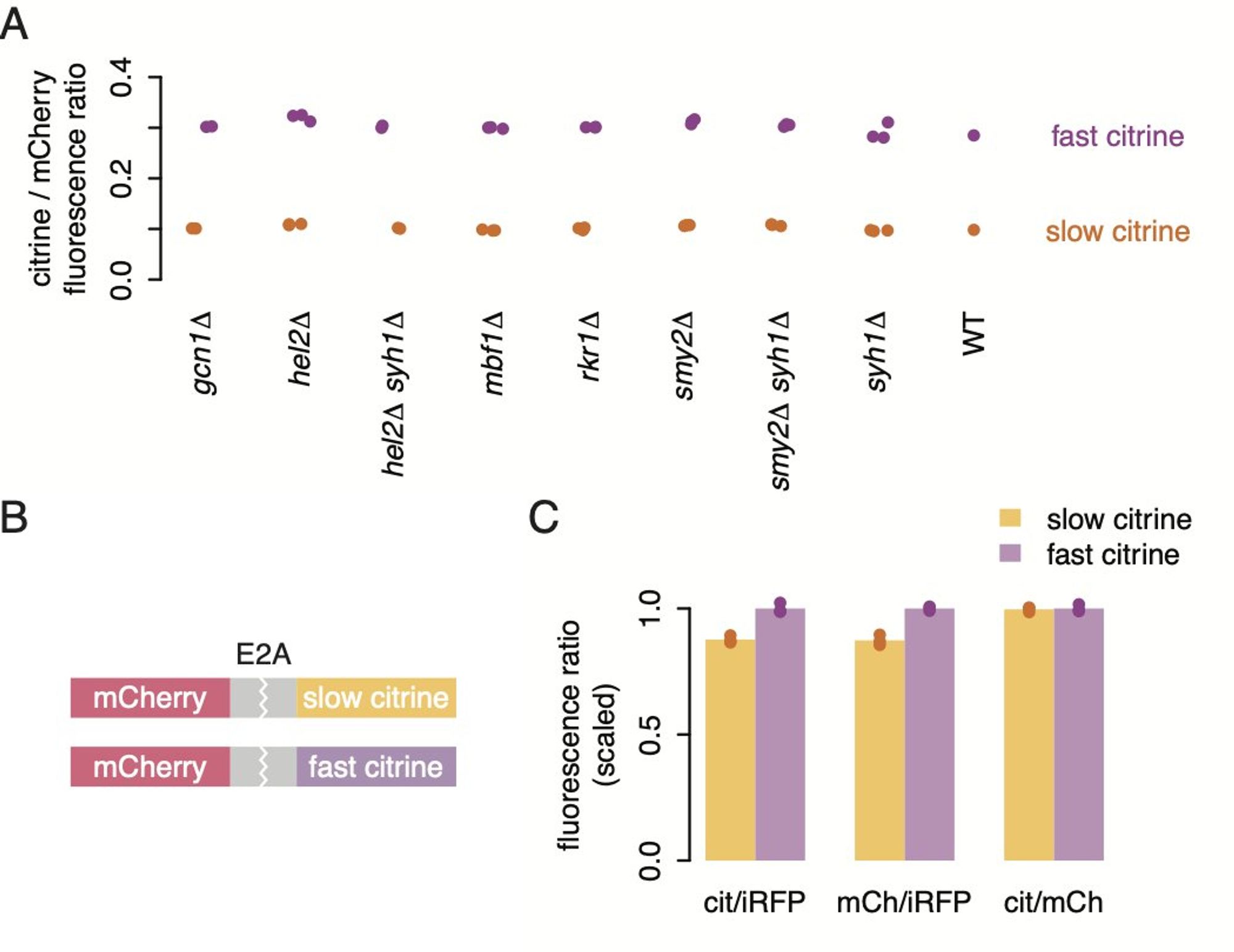
Maybe ribosomes were removed before finishing? We deleted lots of genes that clear up stalled ribosome, with no effect. Maybe slow ribosomes at the beginning got in the way? We shuffled around codons and saw no change.
So we started testing all the ways you could get less protein. A fraction of the effect came from mRNA stability, but the translation efficiency per mRNA was much lower too, on top of that.
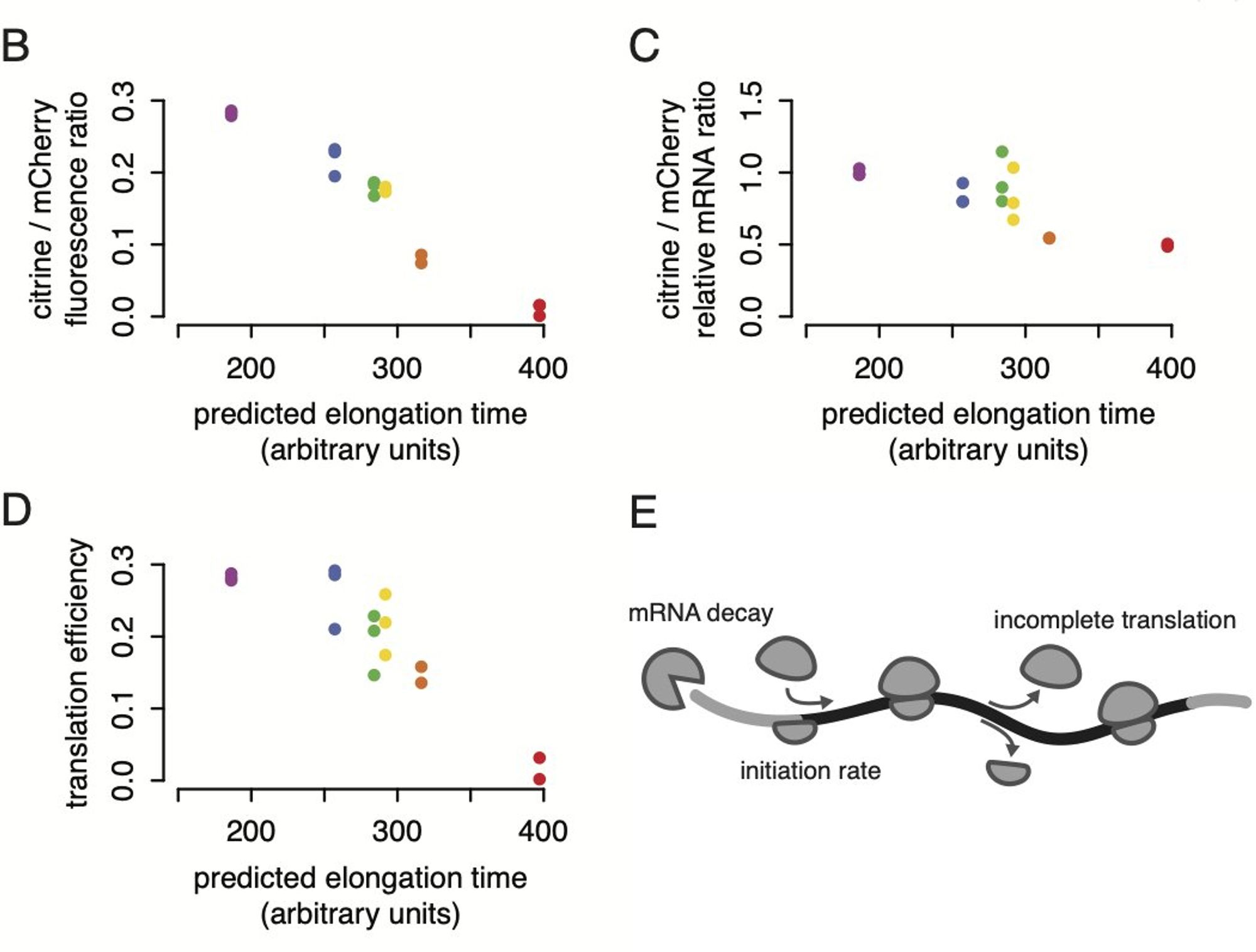
(I’m simplifying of course! You can read the paper, and other great work on this topic from many groups over the years, to see the nuance.)
And if the same number of ribosomes start, but they move slower on the transcript with slow codons, you’d expect a lot more ribosomes on the slow reporter! But we see the same number. (In all figures, purple is the fastest and orange is the second-slowest of our six reporters).