
FoldMason progressively aligns thousands of protein structures in seconds, enabling remote MSA for distant phylogeny. Highlights: structural flexible MSA, LDDT conservation score, friendly webserver 💾 github.com/steineggerla...search.foldseek.com/foldmasonwww.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
Metabuli(분리), our metagenomic read classifier, is now published at Nature Methods. By combining DNA & AA information it recovers 99% and 98% assignments of DNA or AA classifiers. Great work by my student Jaebeom Kim. 📄 www.nature.com/articles/s41...github.com/steineggerla...rdcu.be/dIrgm
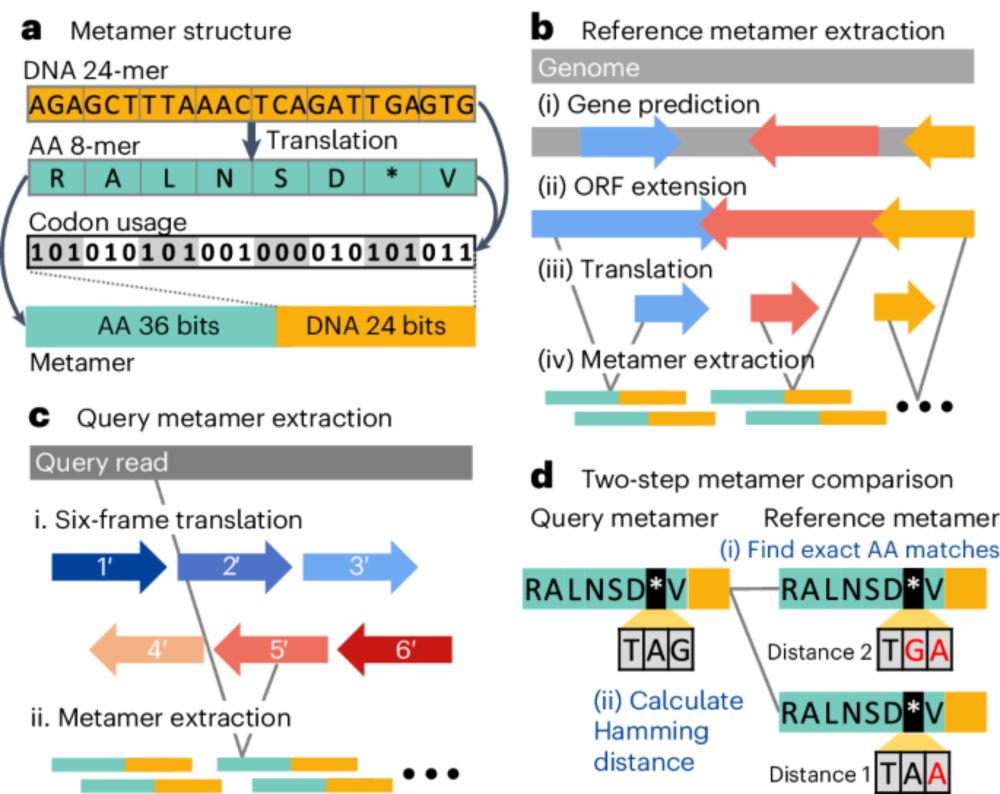
Metabuli enhances metagenomic read classification by jointly analyzing DNA and amino acid sequences for specificity and sensitivity, respectively.
OpenFold has finally been published, in 2024, two and a half years after its initial commit on Github. Once again demonstrating the inanity of peer review in this field www.nature.com/articles/s41...
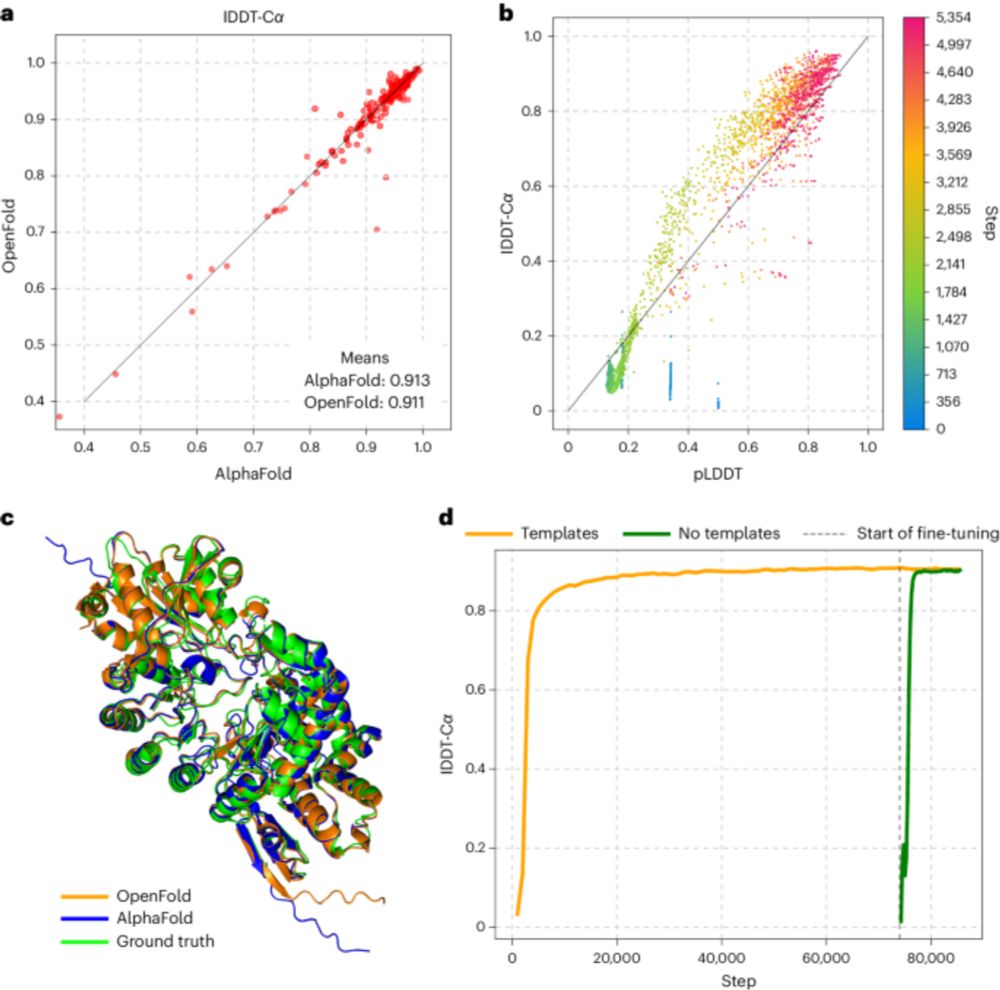
OpenFold is a trainable open-source implementation of AlphaFold2. It is fast and memory efficient, and the code and training data are available under a permissive license.
I think they release af3 once any competitor comes close enough
I reviewed the AlphaFold3 paper from DeepMind for the journal Nature. I tried really hard to get the editors to demand that DeepMind release the code (even an executable) so people could do the many high-throughput studies we saw for AF2 (see image from my review). I failed. So just a server 4 now.

Foldseek-Multimer is a protein complex aligner that is up to 10,000x times faster than SOTA methods without sacrificing quality, enabling the comparison of billions of complex pairs per day. 🧬🧶 📄 www.biorxiv.org/content/10.1...github.com/steineggerla...search.foldseek.com

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
Just 3 years :D
Foldseek got published in 2024 in Volume 42 of Nature Biotechnology. Here is the timeline of FS releases: Source code: 2021/07 Webserver: 2022/01 Preprint: 2022/02 Journal: 2023/05 Print: 2024/02

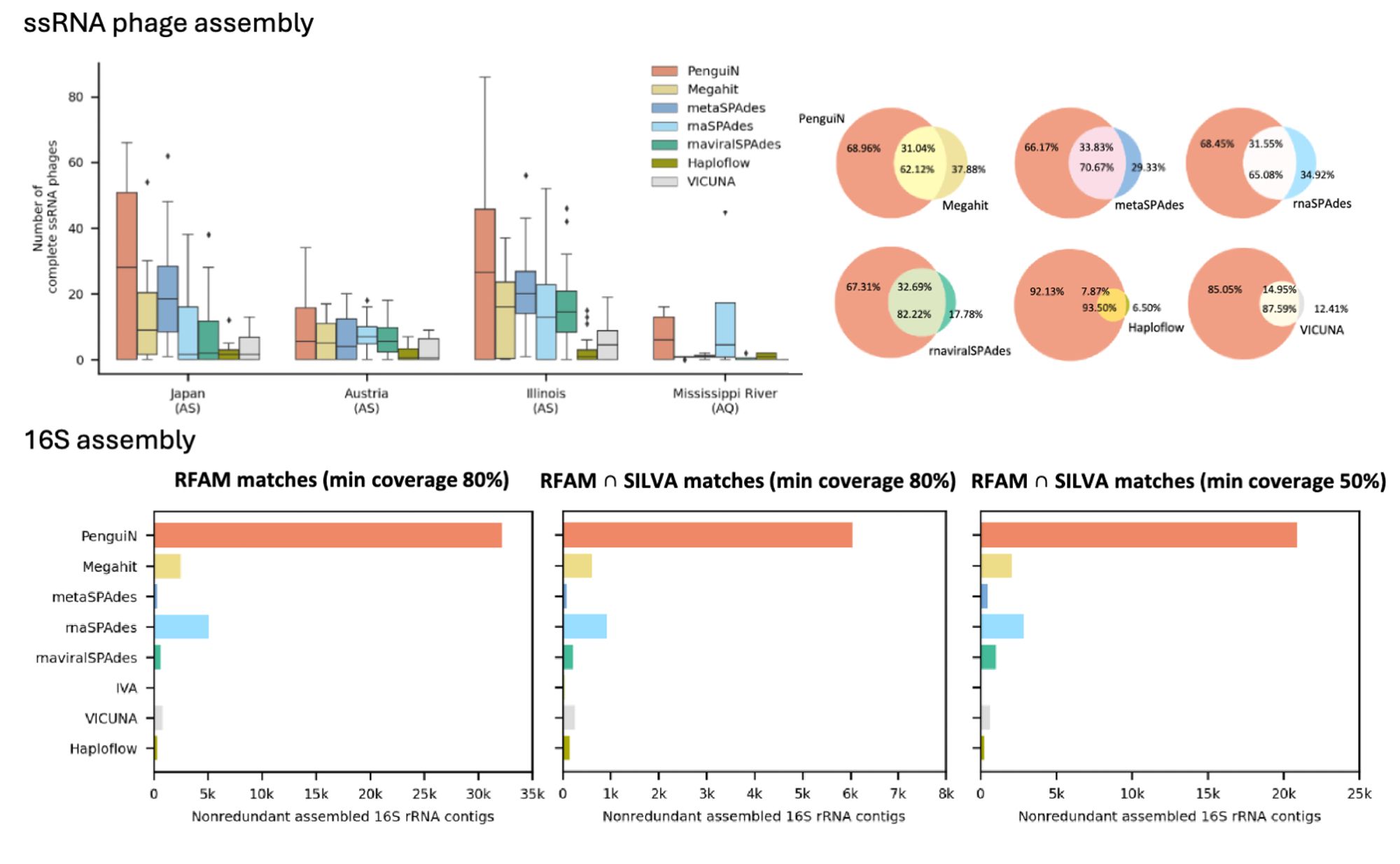
Penguin is our new assembler that reconstructs manyfold more accurate strain-level viral genomes and 16S rRNAs from metagenomes through a novel greedy AA/DNA-hybrid bayesian overlap extension strategy. Work by Annika Jochheim et al. 📄 biorxiv.org/content/10.1...github.com/soedinglab/p...
We released a nice new version of sourmash, along with a super nice new version of the branchwater plugin - faster! lower memory! more convenient! better code! more zhuzh! Hat tip to @bluegenes.bsky.social@luizirber.bsky.social who did all the hard work here!
