Recently out on #bioRxivwww.biorxiv.org/content/10.1... 1/10
Some updated guidance on our gnomAD v4 constraint scores: gnomad.broadinstitute.org/news/2024-03...@gnomad-project.bsky.social team is hard at work on v4.1 and improvements across the board, so expect more updates. Thanks to Katherine Chao for spearheading this blogpost.
Our paper describing a way to infer the phase of rare variant pairs using gnomAD v2 is out now in Nature Genetics. We hope that the resource we generated will be useful when interpreting rare co-occurring variants in the context of recessive disease. www.nature.com/articles/s41...
It’s the final day of giving thanks for the teams that make gnomAD possible. Today is focused on the participants in studies, data contributors (>300!), the Scientific Advisory Board, and our steering committee. 1/6
Day four of giving thanks to the teams that make gnomAD happen is focused on the CNV and SV teams! v4 is the first time we released structural variants at the same time as SNVs/indels, specifically: - CNVs from 464,297 exomes - SVs from 63,046 genomes 1/4

Continuing the week of thanking the teams that make gnomAD possible, today I’m thanking the data generation and operations teams! As a reminder, I'm only highlighting a few individuals of the many that contribute. You can see more on our team page: gnomad.broadinstitute.org/team 1/5

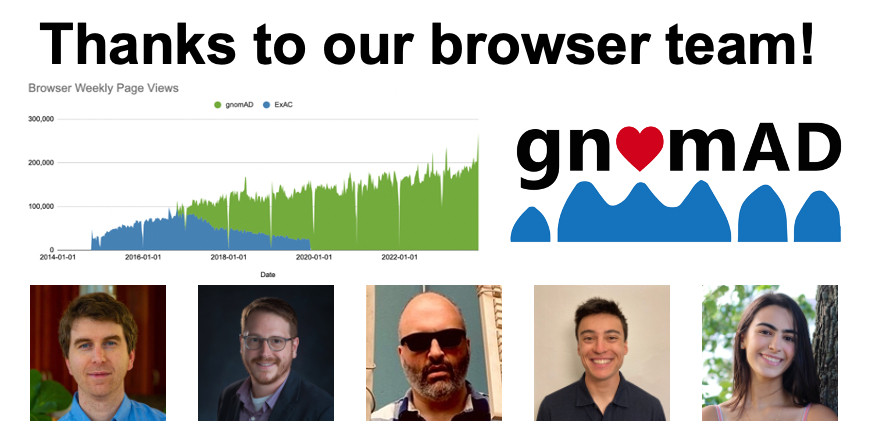
Next up in my week of thanks to the teams that make gnomAD: the gnomAD browser team! With 150k+ views a week, the browser is a crucial part of making gnomAD accessible. Quickly loading data from >800k samples + presenting it in a user-friendly format is no small feat. 1/6
Now that the dust has settled on the gnomAD v4 release, which hopefully many of you have already checked out, I wanted to take this week to thank many of the members of the team who made this possible. First up this week is the amazing production team. 1/7

Helpful description of the choices of population designators in gnomAD: gnomad.broadinstitute.org/news/2023-11...
Constraint scores are now up for v4!