Some updated guidance on our gnomAD v4 constraint scores: gnomad.broadinstitute.org/news/2024-03...@gnomad-project.bsky.social team is hard at work on v4.1 and improvements across the board, so expect more updates. Thanks to Katherine Chao for spearheading this blogpost.
A freely accessible version of the paper can be found here: rdcu.be/dsVXx
Our data are freely available on the gnomAD browser (gnomad.broadinstitute.org@gnomad-project.bsky.social.
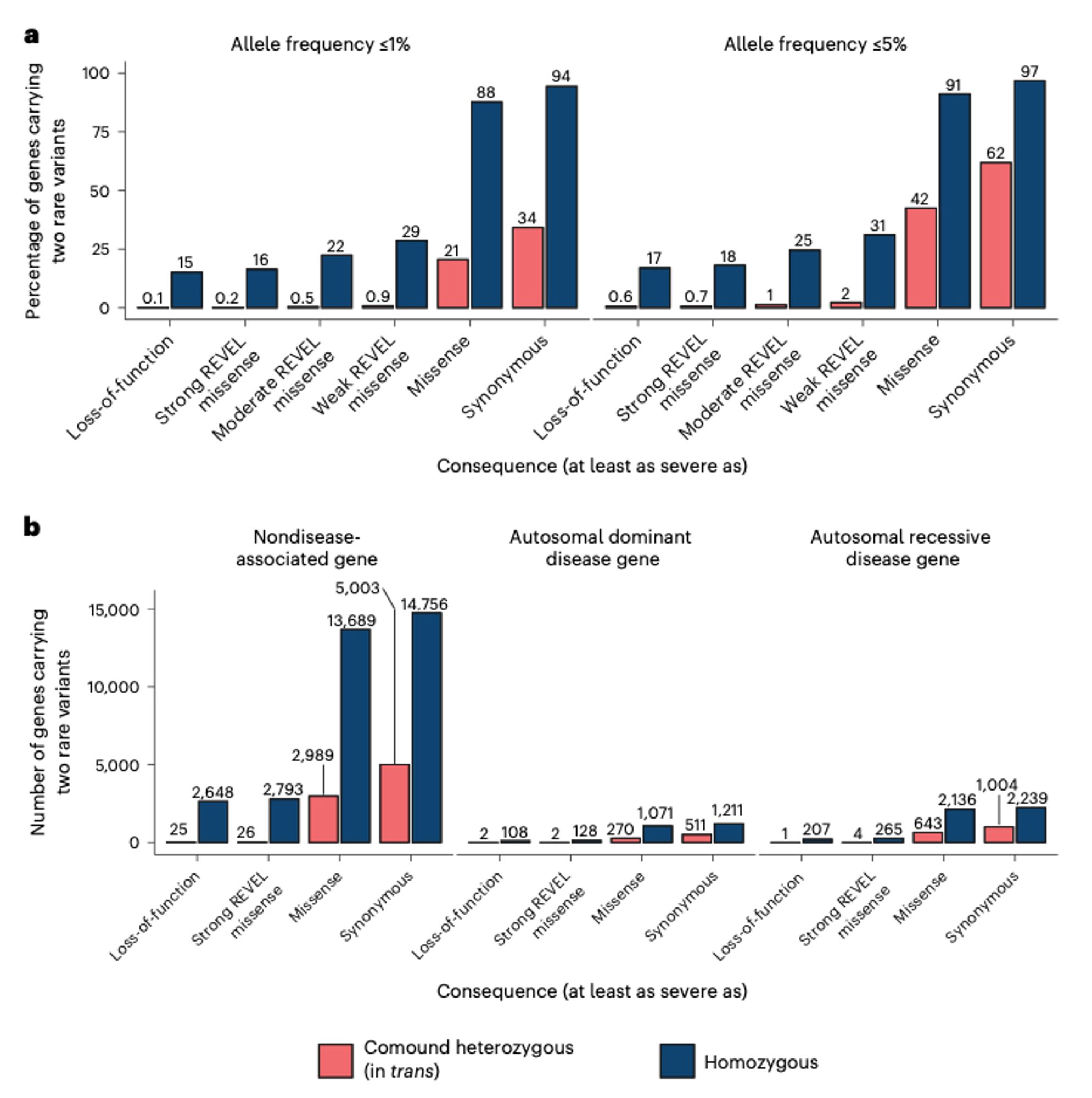
We also counted the number of genes that appeared to have compound heterozygous (in trans) deleterious variants and didn’t find that many. After manual curation, we only found seven genes with high-confidence compound heterozygous loss-of-function variants with some caveats.
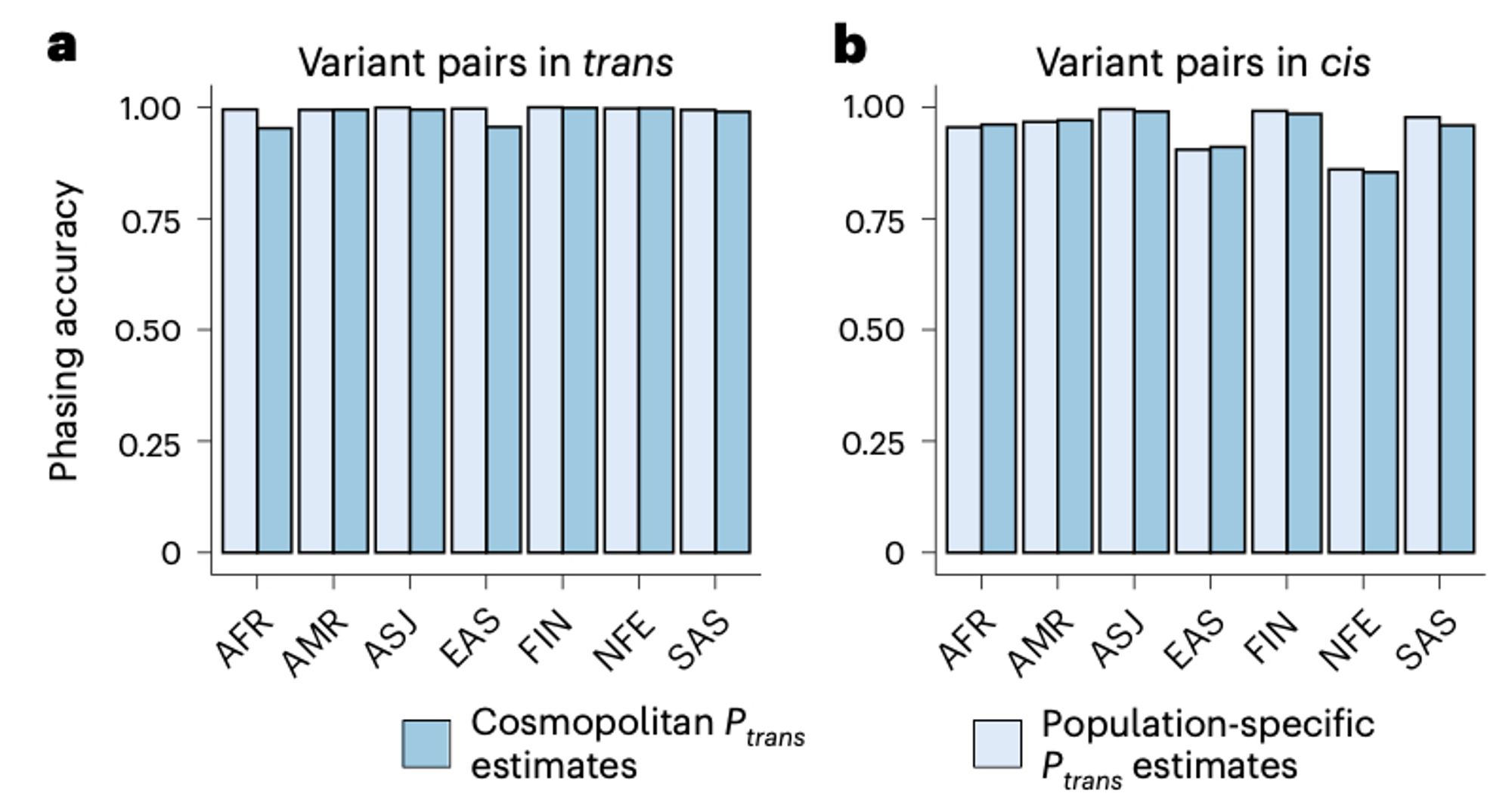
Accuracy of our approach remains high across a range of allele frequencies and across genetic ancestry groups. As expected, genetic distance and highly mutable sites both impact accuracy.
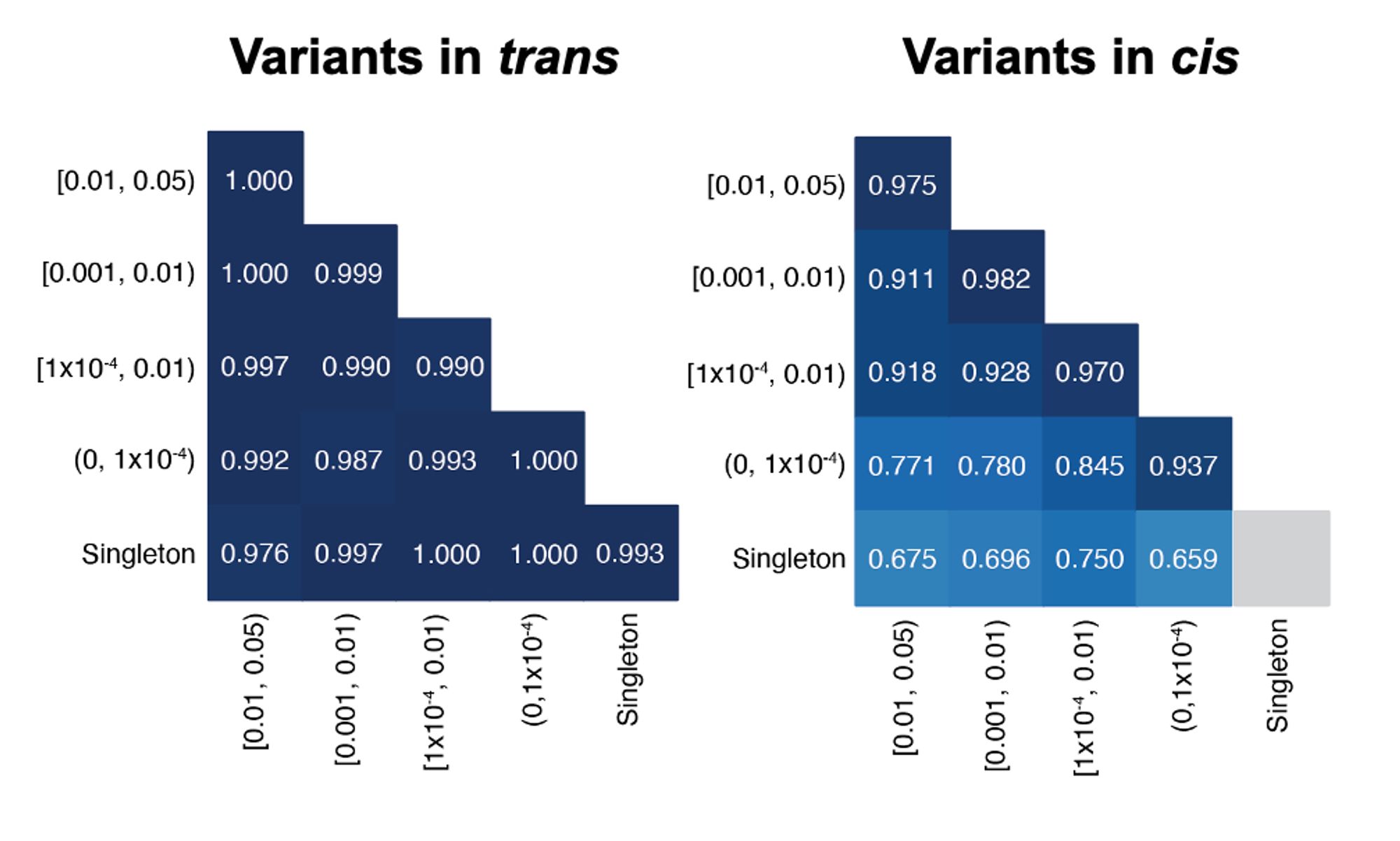
In this work, we used gnomAD v2 to estimate haplotype frequencies, and thereby predict the probability that two rare variants observed in the same gene are in trans. This approach is highly accurate (~96%) for variants with known phase in two independent datasets.
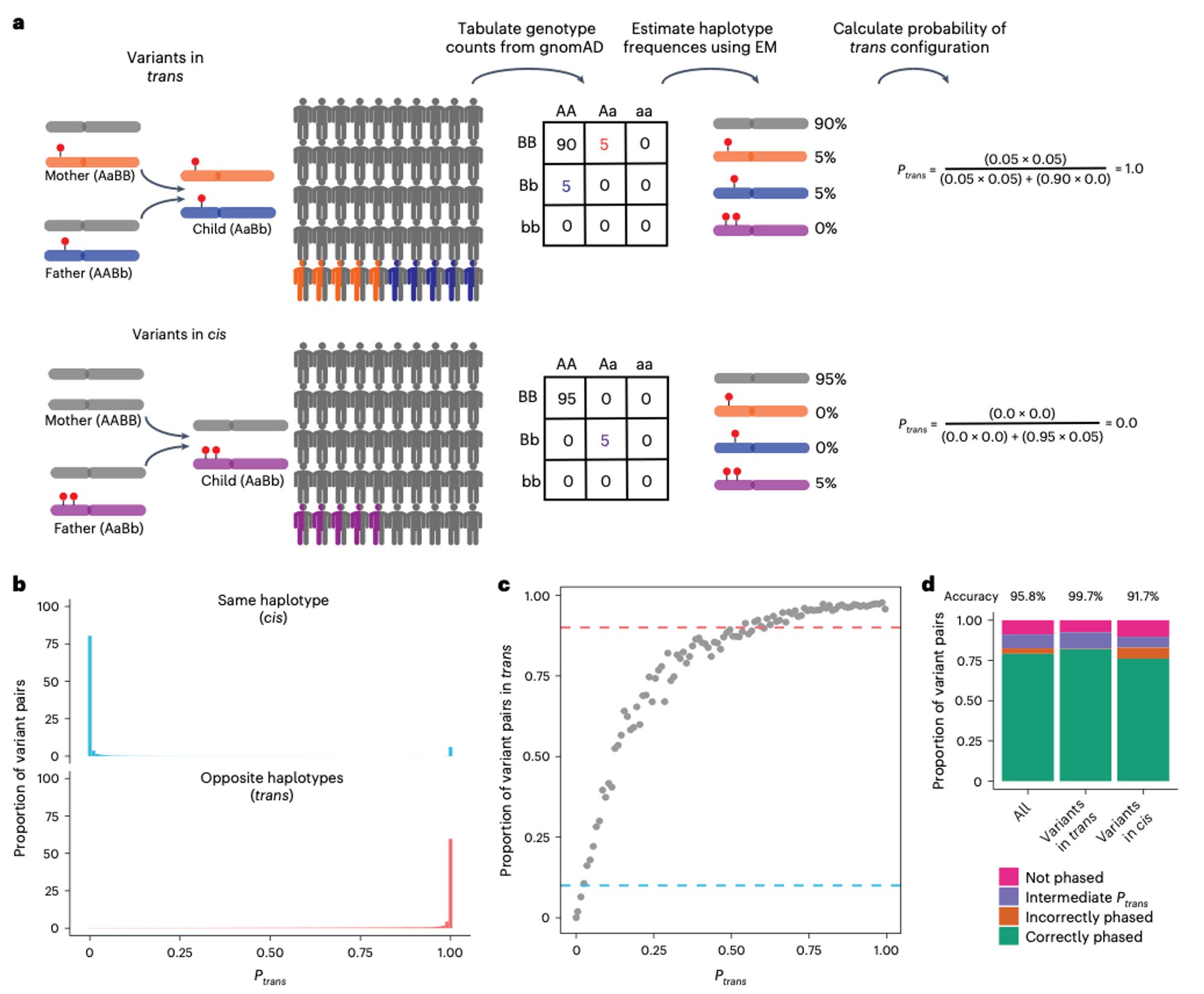
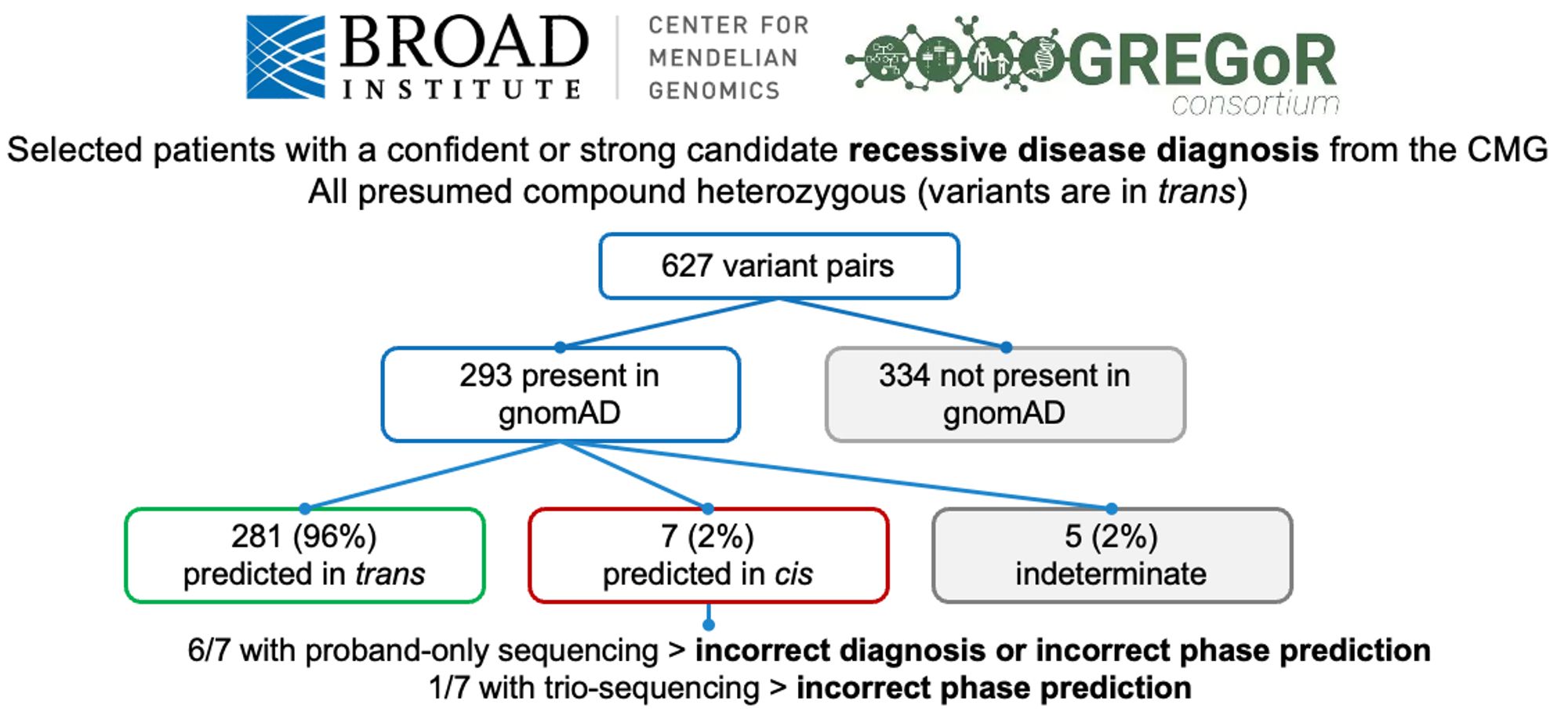
Our paper describing a way to infer the phase of rare variant pairs using gnomAD v2 is out now in Nature Genetics. We hope that the resource we generated will be useful when interpreting rare co-occurring variants in the context of recessive disease. www.nature.com/articles/s41...
As a note, I consulted with a few people this week (Katherine Chao, Sam Baxter, Jack Fu) to make sure that I covered as many of the key players as I could. Any remaining omissions or errors are all my own. 6/6
Finally, I’d like to thank the steering committee of gnomAD, and particularly to our co-Directors: Mark Daly and Heidi Rehm. It’s been fun to be a part of this amazing project and process. 5/6

On the leadership side, we have a wonderful Scientific Advisory Board who provides suggestions and guidance for us. I’m looking forward to more discussions about how we can make this resource better. 4/6
