Whaaat? Are these people nuts there? They let go of the best guy they could have! I guess Universities around the whole world are the same... Such a short-sighted strategy...
Believe it or not, it's the first time I've used it. And the increase in the identified peptides and proteins is independent of the input, from 10 ng down to 32 pg. So I was thinking to just always use it, but... Does it influence the mass spec in the long term in any way? Columns?
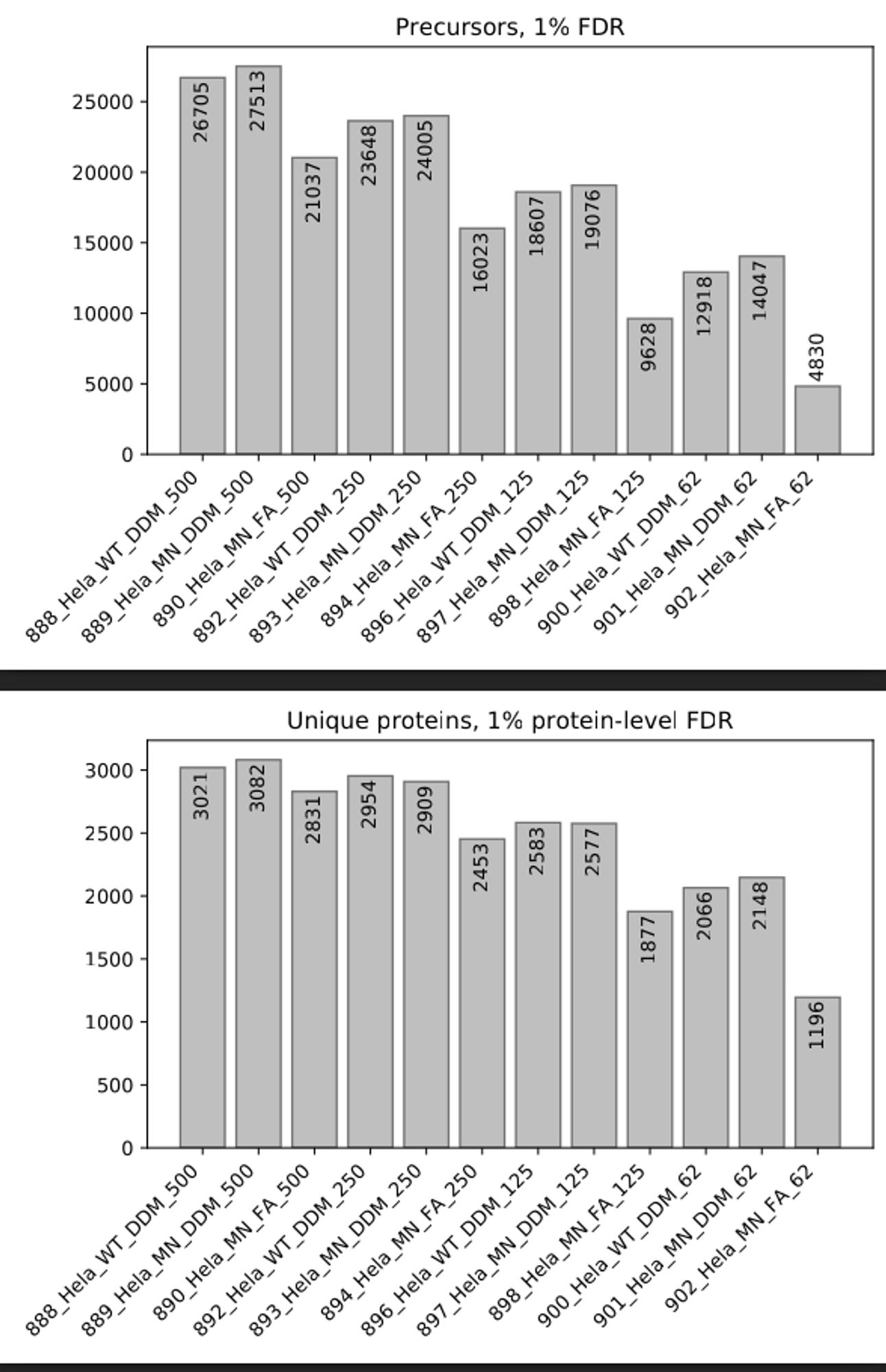
Results from 500 pg down to 62 pg, with DDM and without (samples FA). MN and WT describes different vials used in the autosampler.
In two weeks I have the proper training by Bruker. I want to get my results to them and confront their knowledge and specs of other Ultras.
Yes, I would guess so. I have read recently a paper about this on Orbitraps, and the consensus was the same. Just forgot from whom the paper was ;) Too many things.
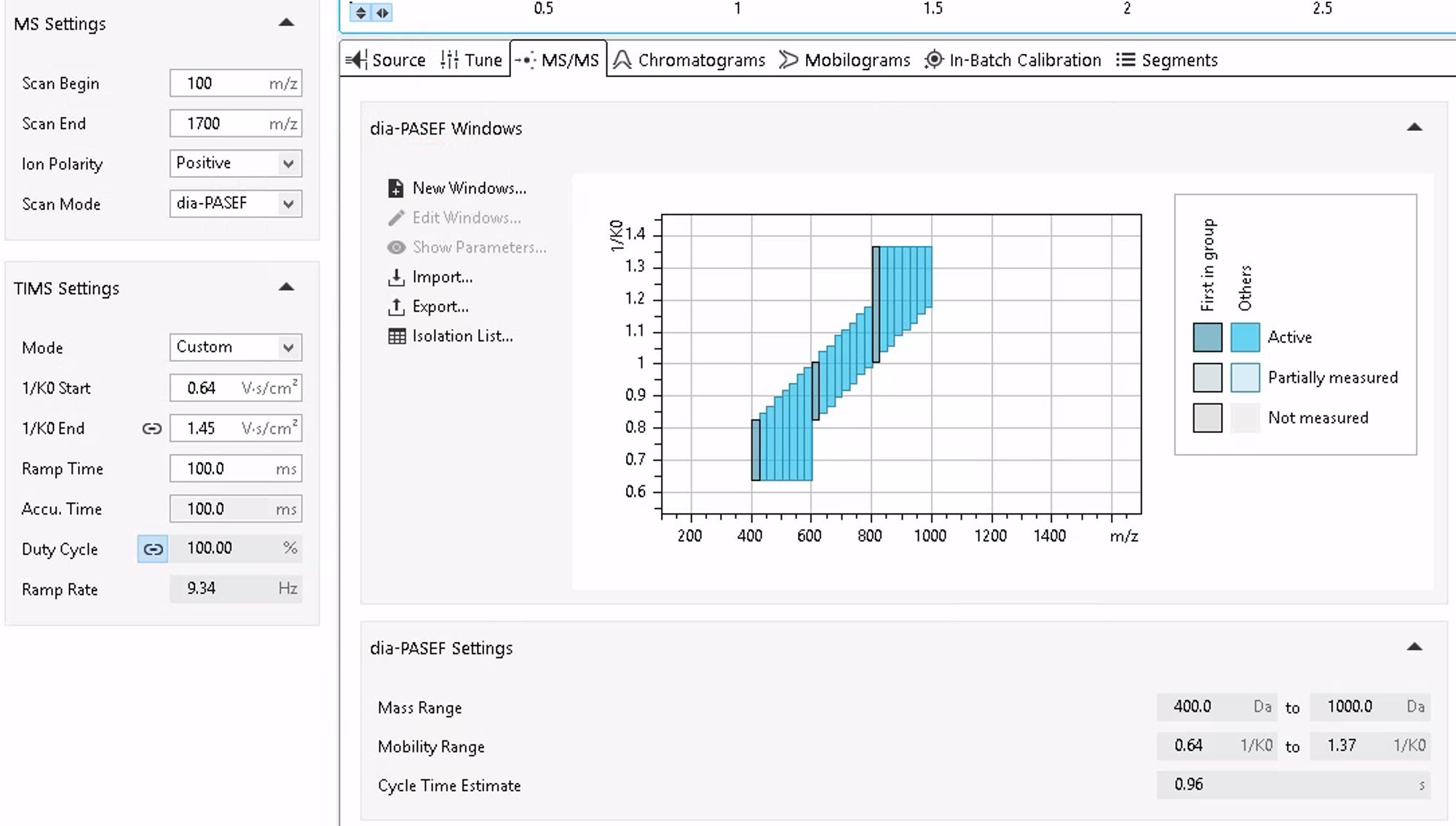
Column: PepSep Ultra It's the default method from Bruker. The only thing I was playing until now with DIA was the ramp time. Faster methods are better (shorter ramp time), regardless of the length of the gradient.
I made more tests with DIA from 100 pg to 150 ng, with gradients from 10 min to 90 min. Anything below 5 ng suffers from gradients above around 20 min. With higher inputs , longer gradients actually give you a tiny little bit more precursors. Full report incoming after I'm back from holidays :)
smh... 🤦♂️
VSCodium

