Have long-read data and want to know what you've really sequenced, and how much of it? Reliable taxonomic labels are often scarce if the target is from a less well-explored group. Fortunately, 2D embeddings from a VAE can tease apart different organisms in read sets: www.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
If you have thoughts on how you would like ModelFinder & PartitionFinder to work in IQ-TREE, I am interested in hearing them. We are re-building this now. Particularly interested in pain points for users! Please repost if you have phylo followers...
To help SMTPB grow & build a community in modeling, theory, simulations, & data analysis in population biology (widely interpreted, incl. ecology, evolution, epidemiology, systematics...), please share this widely, join the listserv (email lbond@stanford.edu), & consider membership (smtpb.org)!
New in Nature - a phylogeny of over 10,000 species (all families, 58% of genera) inferred using Angiosperms353! We've been calling this the "Big Tree" paper for years and I'm glad to see it in print - fantastic work by the Kew PAFTOL team and many others! www.nature.com/articles/s41...
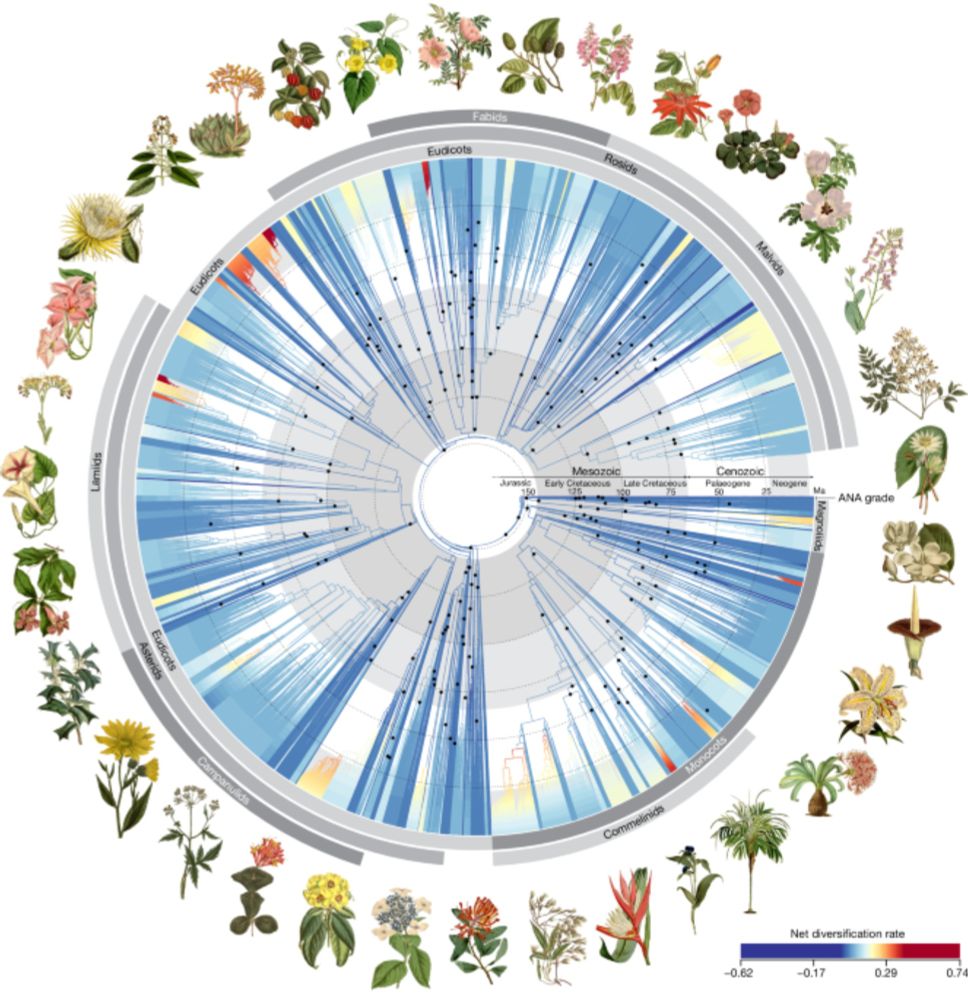
Phylogenomic analysis of 7,923 angiosperm species using a standardized set of 353 nuclear genes produced an angiosperm tree of life dated with 200 fossil calibrations, providing key insights into...
This is something I have been waiting for for a while. A completely novel approach for TE detection. Brilliant!
Identification of transposable element families from pangenome polymorphisms https://www.biorxiv.org/content/10.1101/2024.04.05.588311v1

Background: Transposable Elements (TEs) are fragments of DNA, typically a few hundred base pairs up
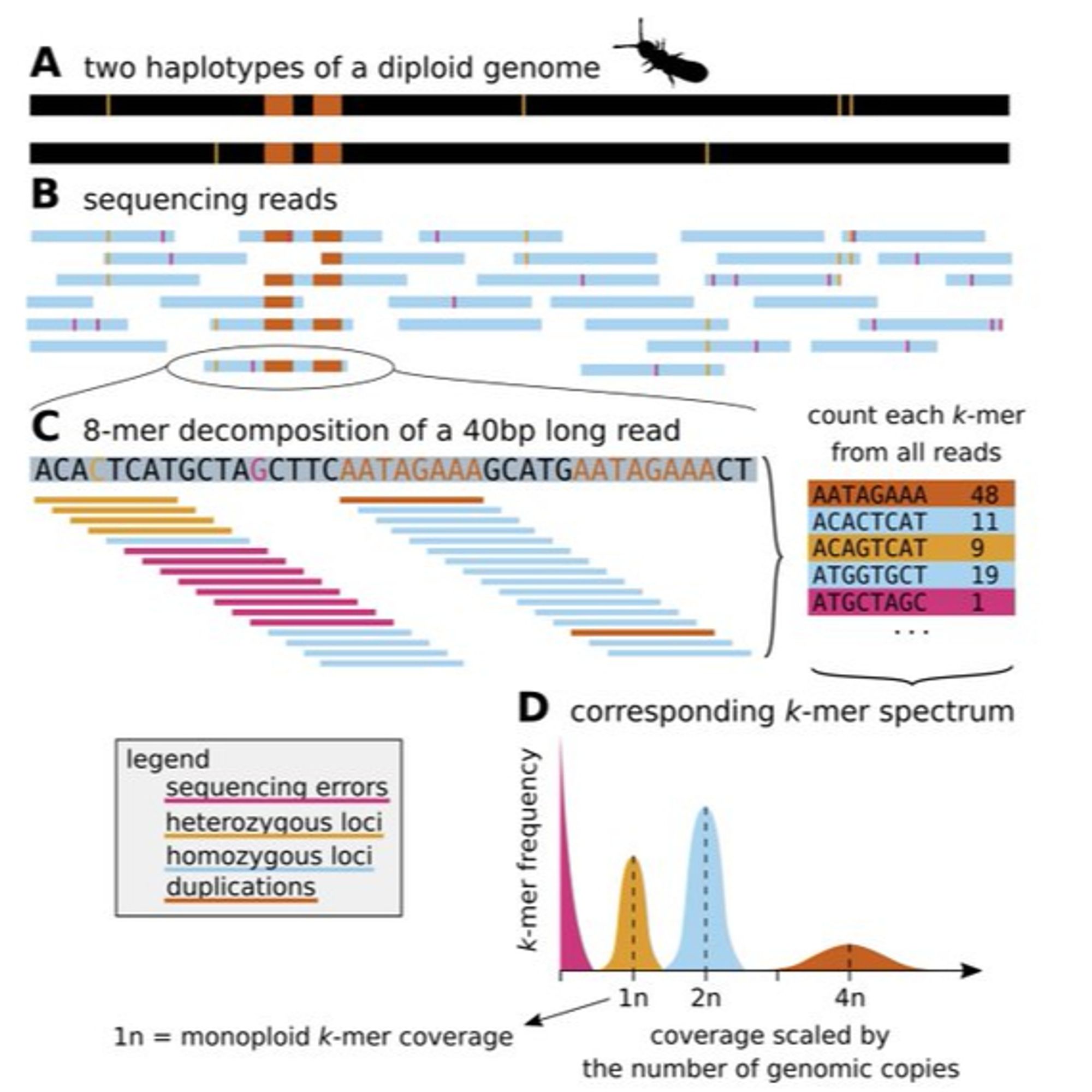
I am delighted to share a preprint of our *Guide to k-mer approaches for genomics across the tree of life* By Katie Jenike, Lucia Campos-Domínguez, Marilou Boddé, José Cerca, Christina N. Hodson, @mikeschatz.bsky.socialarxiv.org/abs/2404.01519
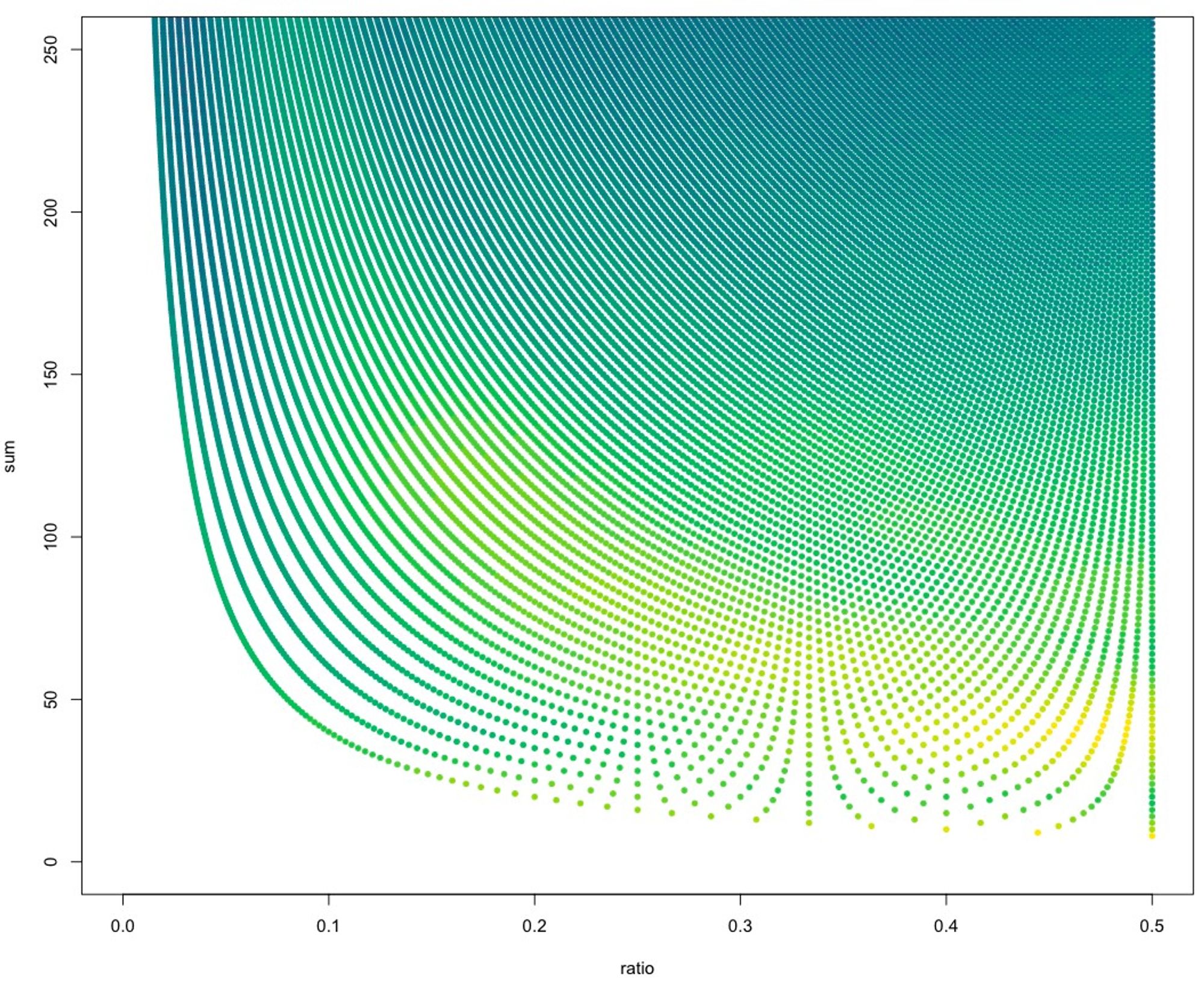
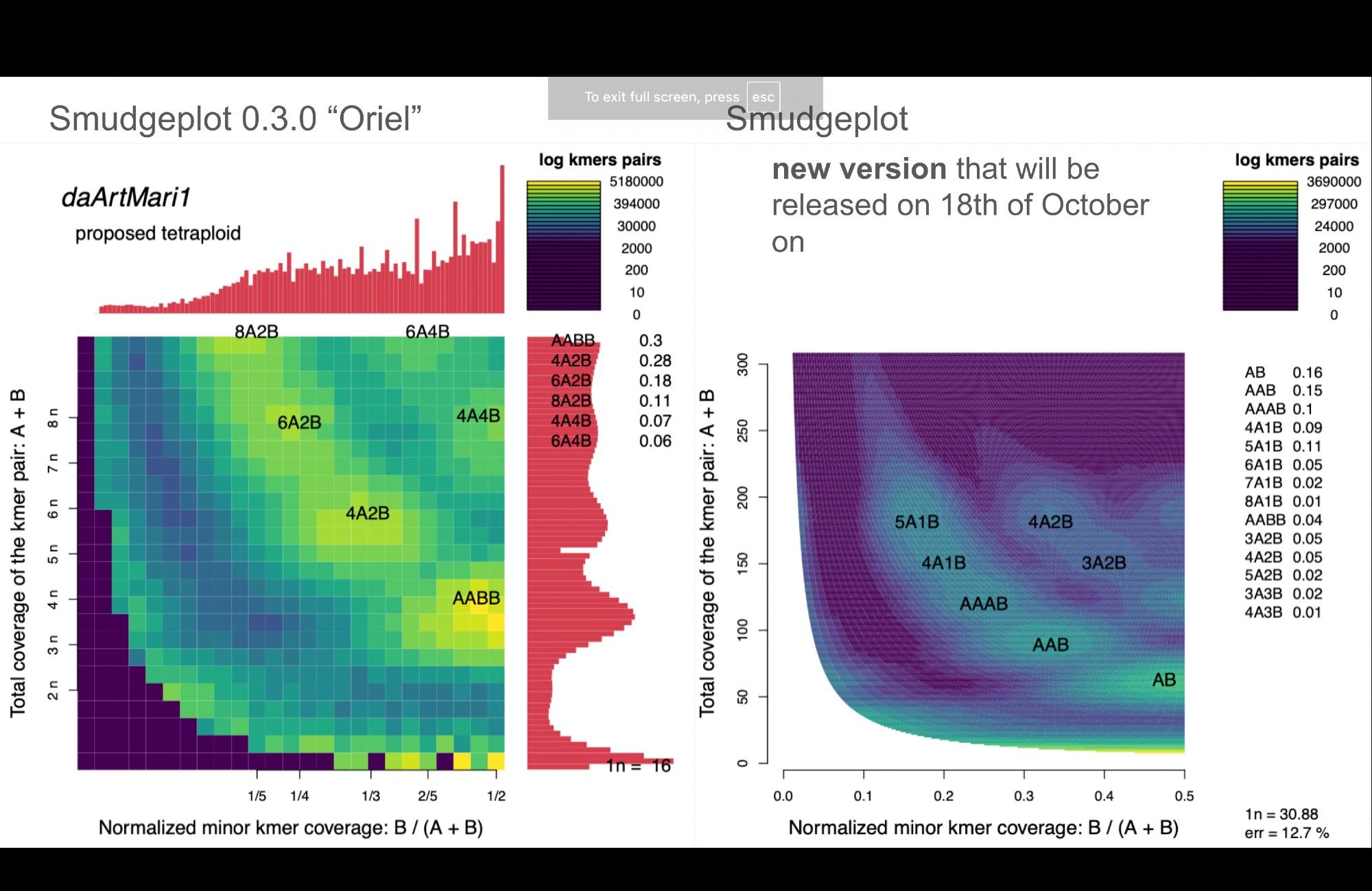
Did you miss #smudgeplot on social media? We are steadily progressing in development of the new version with lots and lots new things in there!