
Authors - post a description of the software, Nature - write a news feature covering this… dressing it up with the veneer of a peer reviewed article is a damaging farce. Submit for peer review when you’re ACTUALLY READY to be reviewed.
Fundamentally, if the journal is not actually peer reviewing the software, what is the point of them publishing an article describing it? Answer: Advertising (for the authors) and citations (for the journal). None of this lives up to the purpose or standards of scholarly publishing.
I'm going to translate their editorial into English: It's important to note that we at Nature have really high standards for materials, data, and code sharing, which we will enforce assiduously for plebes. But do remember that while all authors are equal, some are more equal than others.

If you know a lot about AMR mechanisms in a bacterial pathogen, consider joining us in ESGEM-AMR to share your expertise and develop #AMRrulesbit.ly/AMRrules

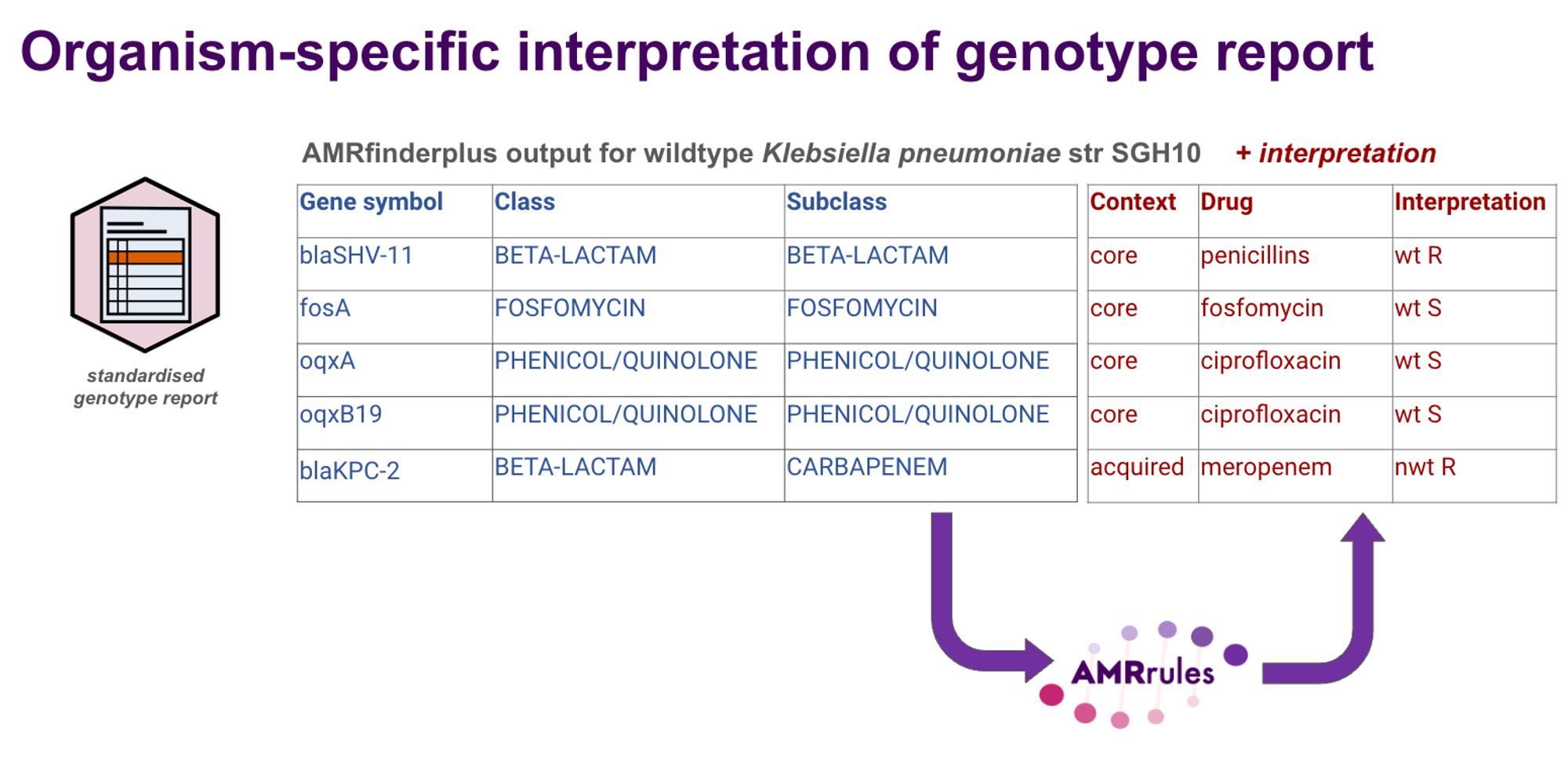
ESGEM, the ESCMID Study Group on Epidemiological Markers is launching a working group to develop standards for interpreting AMR genotypes. Much like EUCAST guidance for interpreting lab phenotypes as S/I/R, we want to define the AMRrules for interpreting genotypes.
Very excited to announce our transdisciplinary AMR workshop 🎉 Join us to hear perspectives and discussion from patient activists, journalists, philosophers, microbiologists, historians, clinicians, policy makers, and artists. More info: www.torch.ox.ac.uk/event/commun...

Our dept at Trinity College Dublin is looking for a Tenure-Track Assistant Professor! Details below 👇 www.jobs.ac.uk/job/DGO006/a...
New preprint out now - stable bacterial strain nomenclature with Life Identification Numbers using a gene-by-gene approach. From our wonderful collaborators at the Institut Pasteur. www.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
How does the ONT R.10.4.1 yield compare with R.9.4.1. I remember seeing some data from ages ago that showed that R.10 yield was lower. Still true?
Would it be great to have a tool that harmonises all the different AMR prediction tool outputs? We agree and outline it in this paper! www.biorxiv.org/content/10.1...

bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
