PDB depositors provided an enormous wealth of open access 3D structure data that helped drive breakthroughs in protein structure prediction More at Open-access data: A cornerstone for artificial intelligence approaches to protein structure prediction www.cell.com/structu...
Delighted are we too!! Our studies on lncRNA actions on gene regulation in pan-cancer system.... 1.RNPS1 among others: doi.org/10.7717/peer... 2. MALAT1 among others:doi.org/10.1038/s41598-022-11214-8 3. MALAT1-RNPS1 complex in alternative splicing:dx.doi.org/10.1021/acsomega.4c04467
Victor Ambros and Gary Ruvkun get the Nobel Prize for microRNAs! Such an incredibly well-deserved win!
The Nobel Prize in Physiology or Medicine 2024 was awarded jointly to Victor Ambros and Gary Ruvkun "for the discovery of microRNA and its role in post-transcriptional gene regulation."
New function for an 'old' chromatin remodeler: great collaboration with @nitikataneja.bsky.social@nhoitsma.bsky.socialwww.biorxiv.org/content/10.1...
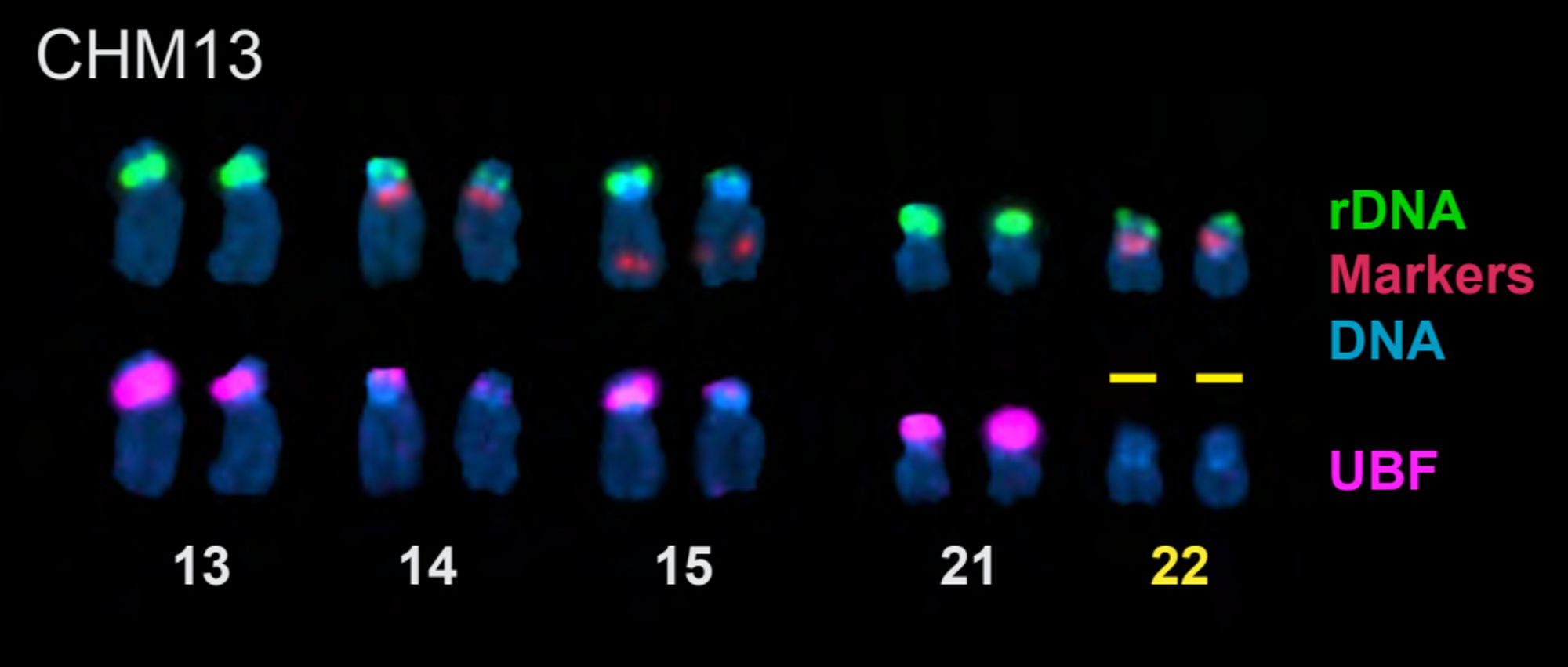
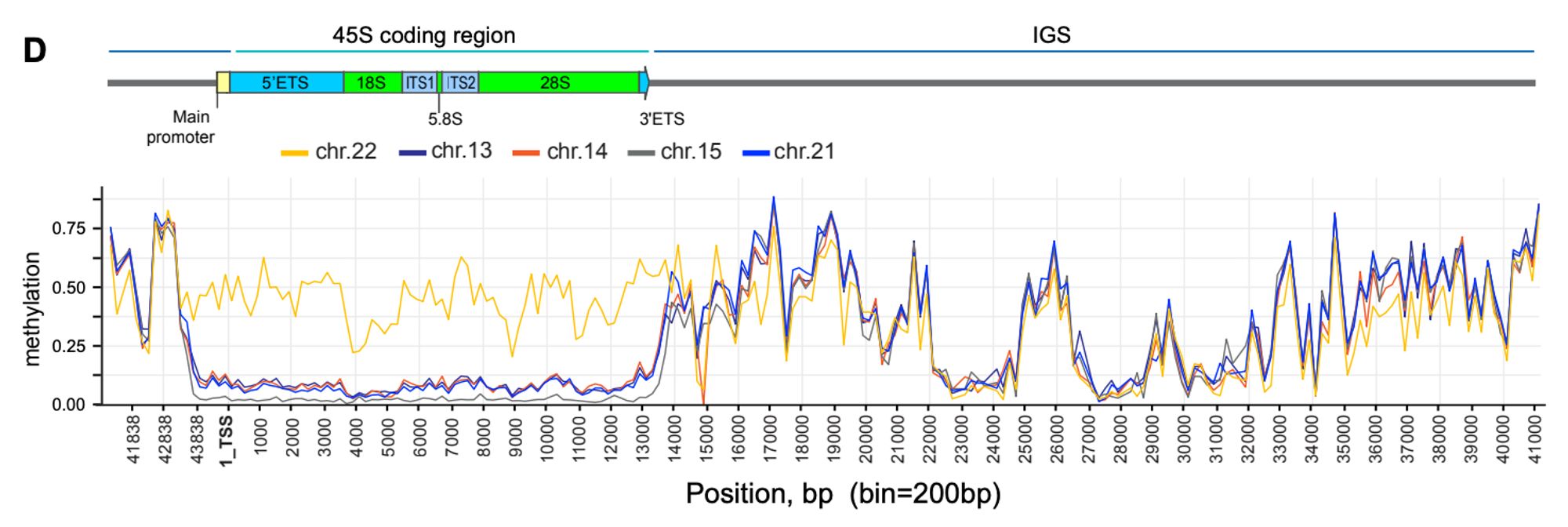
"Epigenetic control and inheritance of rDNA arrays" New preprint from the T2T acrocentrics team! We find a tight link between methylation and transcription of the rDNAs, including the occasional silencing of entire arrays www.biorxiv.org/content/10.1...
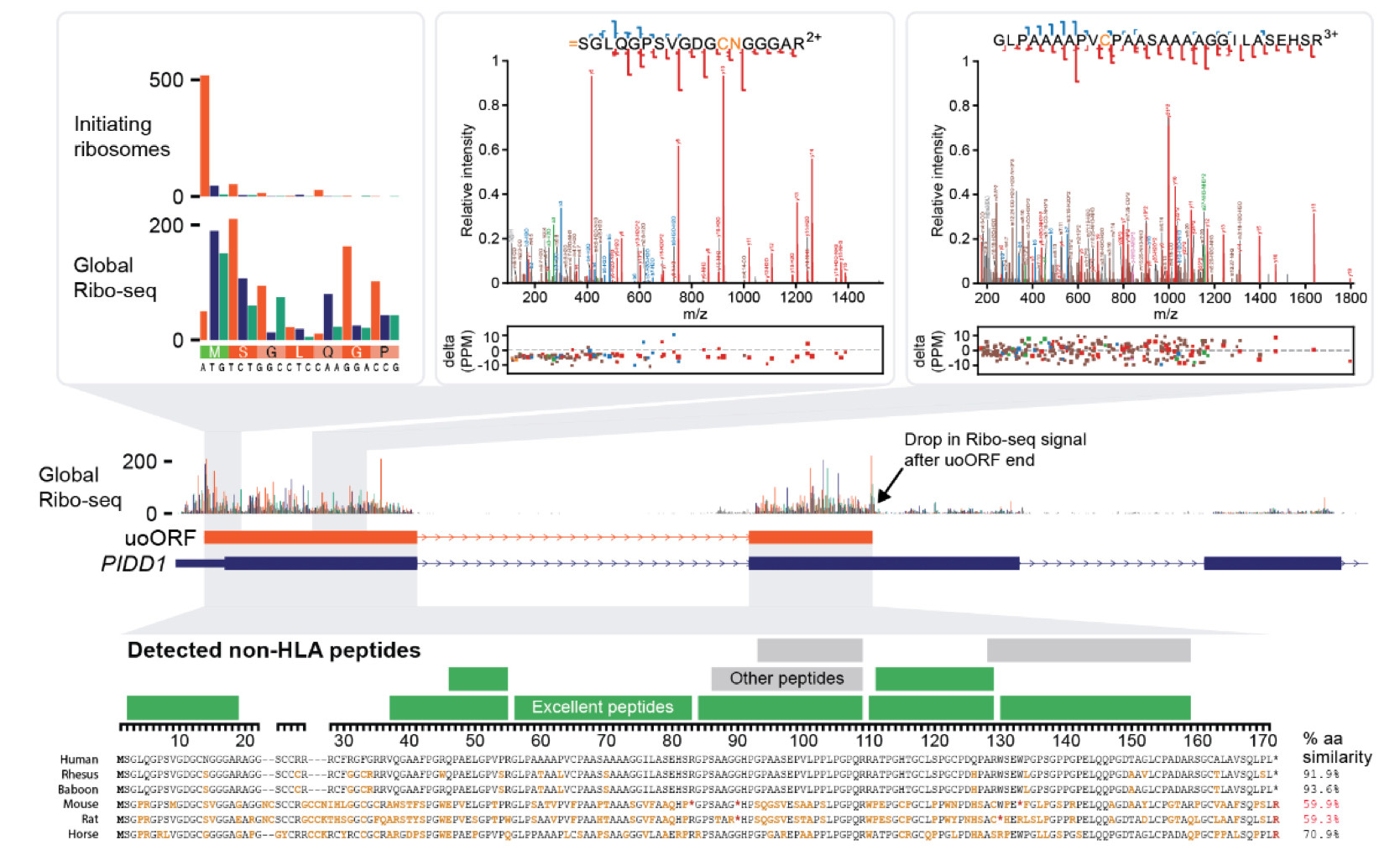
5/ This work is a definitive statement on the capacity for small ORFs to produce the highest quality evidence in mass spectrometry data. In collaboration with GENCODE and PeptideAtlas, these ORFs and peptides will be supported for public view and public research resources.
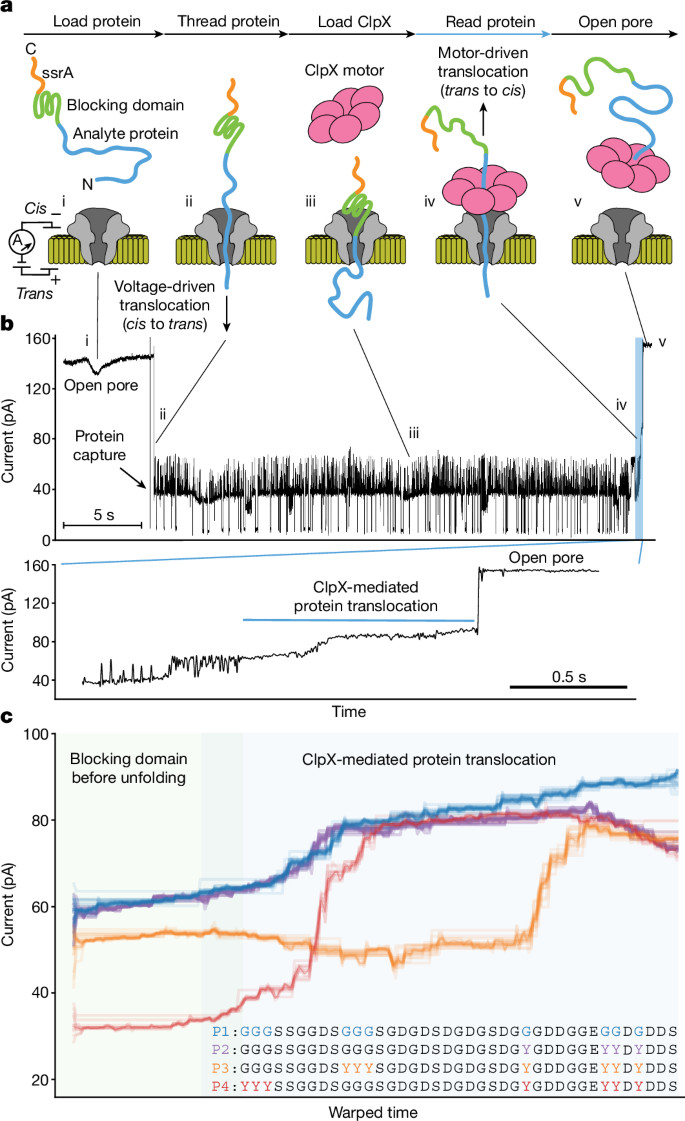
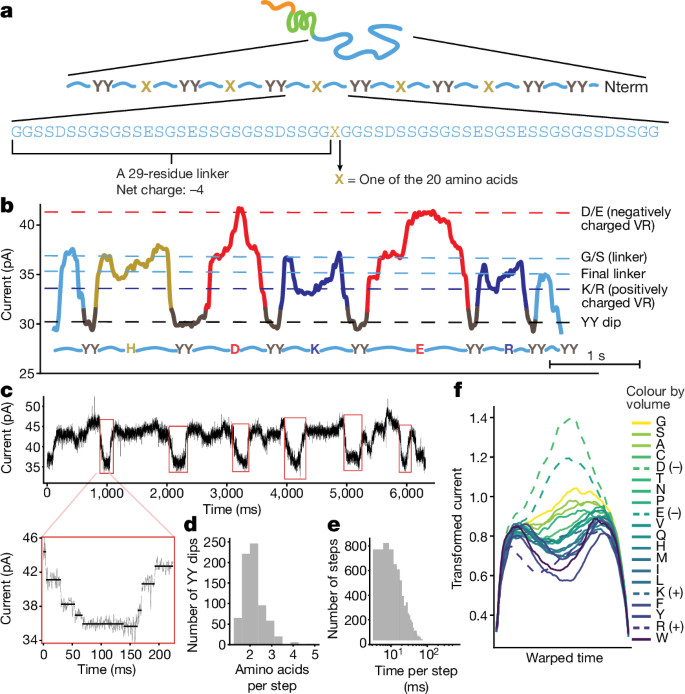
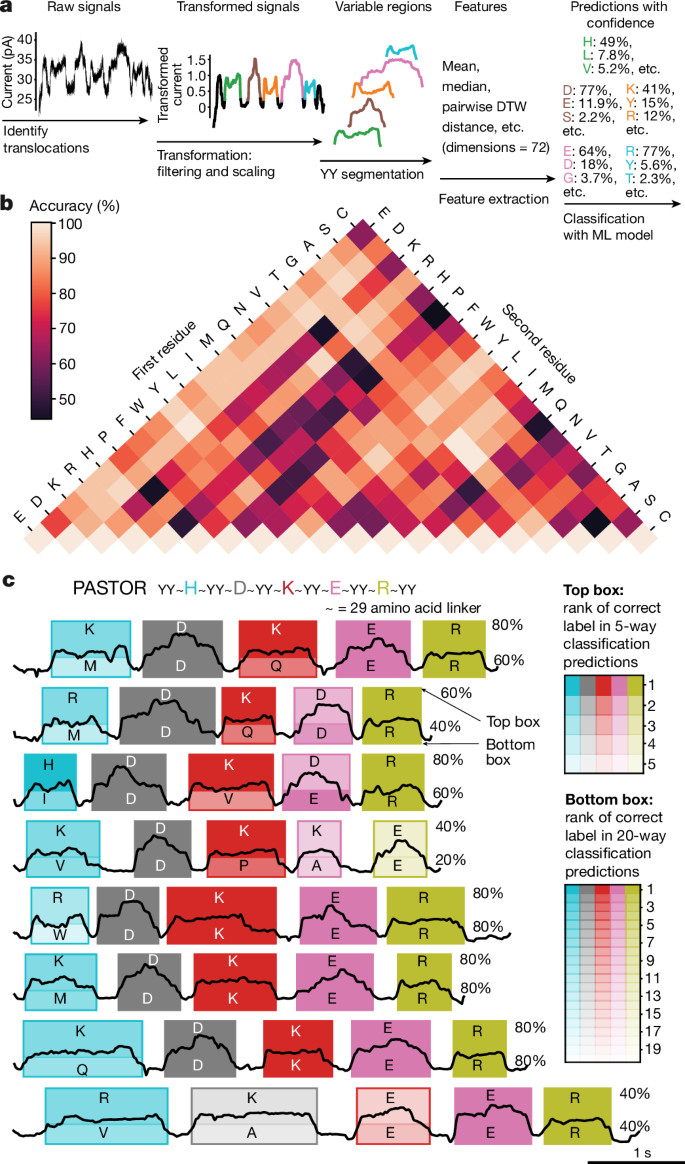
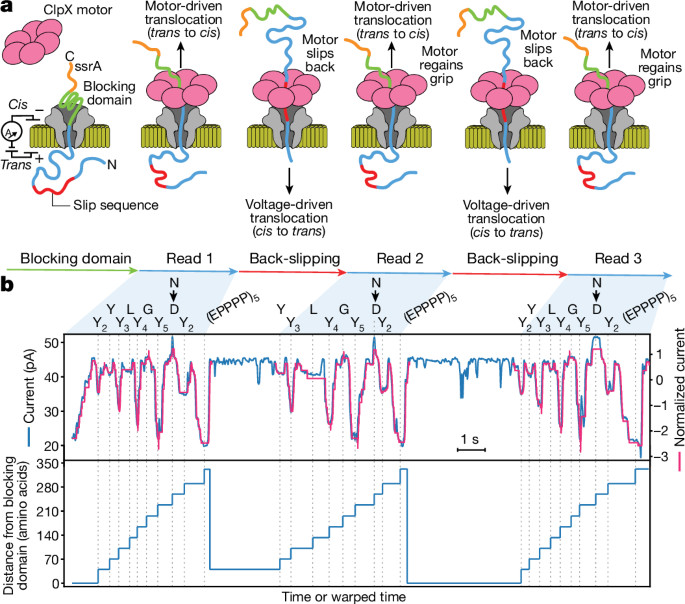
Multi-pass, single-molecule nanopore reading of long protein strands https://www.nature.com/articles/s41586-024-07935-7https://github.com/uwmisl/PASTOR-sequencing
Is there a way to change font styles and make the interface far more professional or cool?
Tried BIMAS, SYFPEITHI, IEDB, MHC motif atlas?
Pollex, T., Marco-Ferreres, R., Ciglar, L., Ghavi-Helm, Y., Rabinowitz, A., Viales, R. R., Schaub, C., Jankowski, A., Girardot, C., Furlong, E. E. M. (2023).. Chromatin gene-gene loops support the cross-regulation of genes with related function. Mol Cell. www.cell.com/molecular-ce...
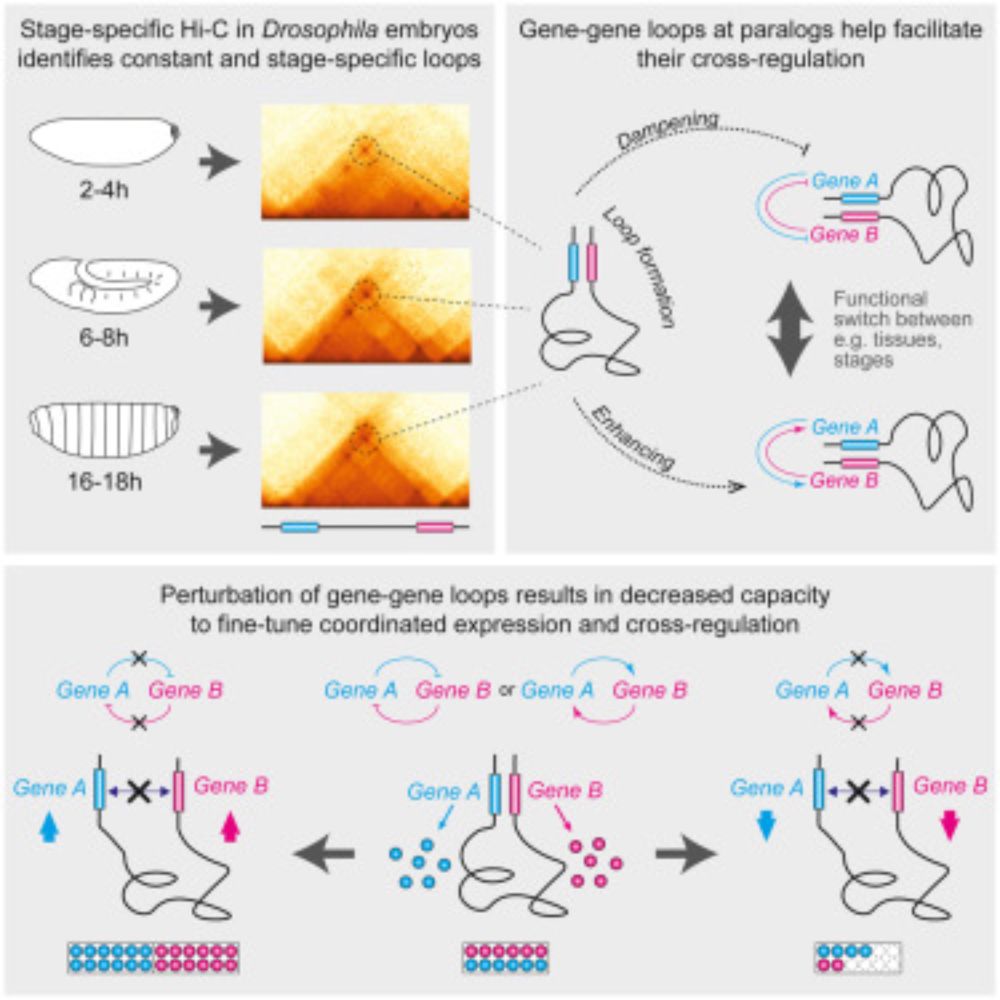
Genes with related function often have co-regulated expression. Pollex et al. show that many paralogous genes form a chromatin loop that sustains their co-expression. Breaking the loop increases or de...