We focused on peptidoglycan hydrolases, enzymes that cleave peptidoglycan. Archaea don’t have peptidoglycan (some methanogens have something similar but different, called pseudomurein). So they should have no cell-intrinsic use for peptidoglycan hydrolases, right? 6/10
I love Allbirds and what they’re trying to do in the fashion space, but the regenerative wool claims are likely based on some flimsy accounting. @metaomicsnerd.bsky.socialwww.fastcompany.com/91185280/all...

Unlike many of its peers, Allbirds has moved beyond deceptive net-zero claims.
There are differences in which methanogens associate with protozoans in the rumen. For example, this study found that Methanobrevibacter spp. - a particularly important genus - were more likely to be associated with protozoa than live freely www.microbiologyresearch.org/content/jour... 13/?

Structures of free-living and protozoa-associated methanogen (PAM) communities from forage-fed cattle were investigated by comparative sequence analysis of 16S rRNA and methyl coenzyme M reductase (mc...
Getting back to protozoa, methanogens have a weirder relationship with them. Methanogens can stick on the outside of protozoa, sure. But methanogens also often live INSIDE protozoa. 12/?
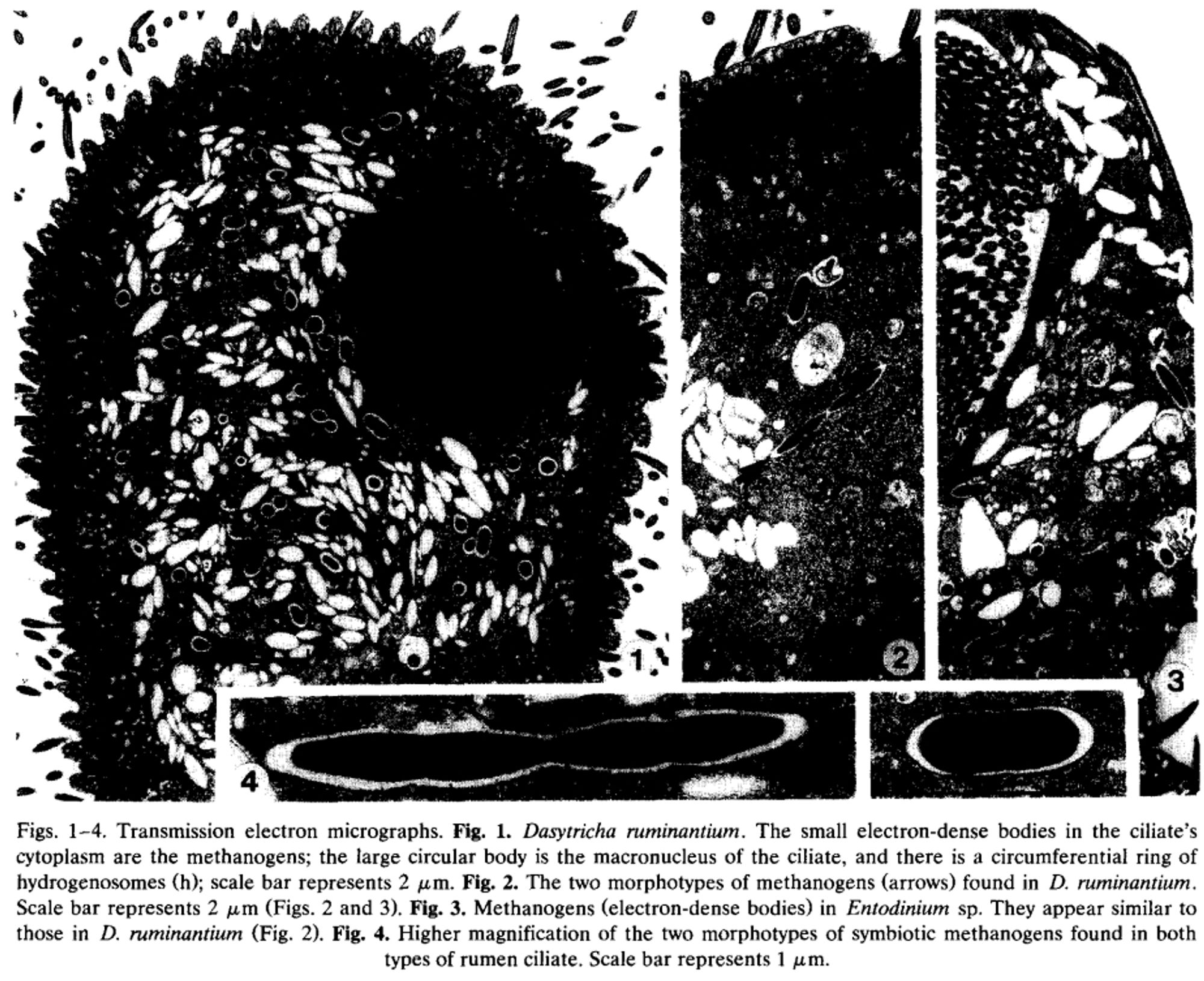
Methanogens have evolved a couple of really weird ways to make sure they acquire hydrogen. The first, which is a bit more broadly applied, is the use of adhesin-like proteins (ALPs) to stick onto the surfaces of hydrogen producers. This is a strategy they employ with fungi. 11/?
This energy, of course, comes in the form of fermentation products of sugar, which comes from plants, which eat the sun. So methanogens have to get by with a little help from their friends, in the rumen. In academic terms, we call this syntrophy. 10/?
Protozoa, much like fungi, have close relationships with methanogens in the rumen. Methanogens, as you remember, are the archaea in the rumen and the producers of methane, which comes out in the burps that are killing us all. 8/?
Targeted isolation of Methanobrevibacter strains from fecal samples expands the cultivated human archaeome Nice paper isolating methanogens using syntrophic bacteria. Also turns out you can screen human breath for methane to identify people with more gut methanogens! www.nature.com/articles/s41...
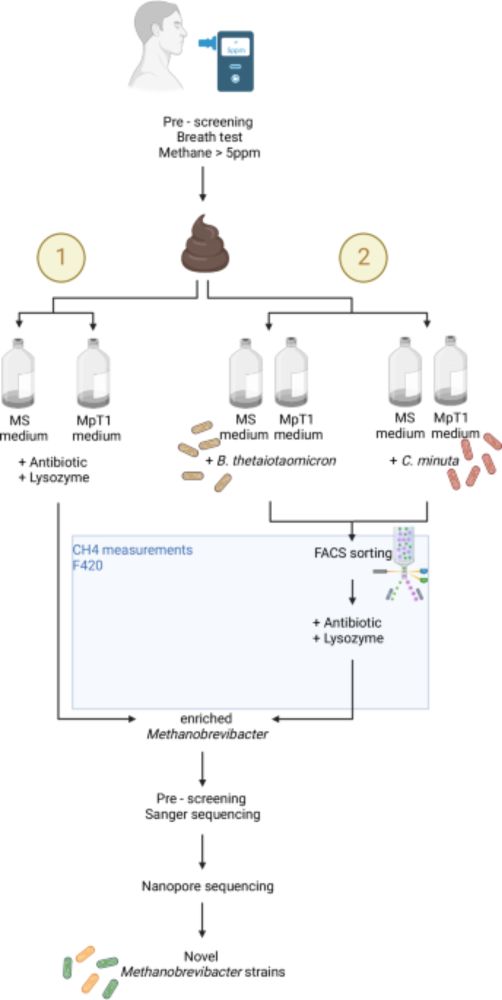
Here, the authors present a new method for isolating methanogenic archaea from human fecal samples and establish stable archaeal cultures yielding nine previously uncultivated strains, which upon furt...
The main challenge in methane emissions reduction is how to reduce rumen acidity. When methanogens are inhibited, most hydrogen leaves the rumen, but some of it stays behind. We're working on how to direct it into organic acids that the cow absorbs - that's the current research frontier. 12/12
Enter the methanogens! Most methanogens in the rumen are hydrogenotrophic methanogens, which means they reduce CO2 to methane using hydrogen or formate. This takes up all the excess hydrogen and formate. Methane doesn't go into solution, but instead leaves the animal. 11/?