for their binding sites. Anyway, this is not a serious test. We'll need the code to go large-scale. It's just what I came up with given the limit of 10 jobs/day. After this limited experience with the webserver, I'm impressed! 🤯 END
also the others we have at least one perfect solution in 6/8 cases. I also tried with totally random DNA. As I expected it binds DNA the normal way (helices in groove) even if there's no LexA binding site. Not necessarily a "wrong" prediction given how bacterial TFs "search" 8/
Case 3 is particularly convincing because the site came out quite different from the consensus. It's not the classical CTGT-n8-ACAG, but AF3 predicts that the dimer binds there. AF3 can propose more than one model, but I was only looking at the first proposed. If I consider 7/

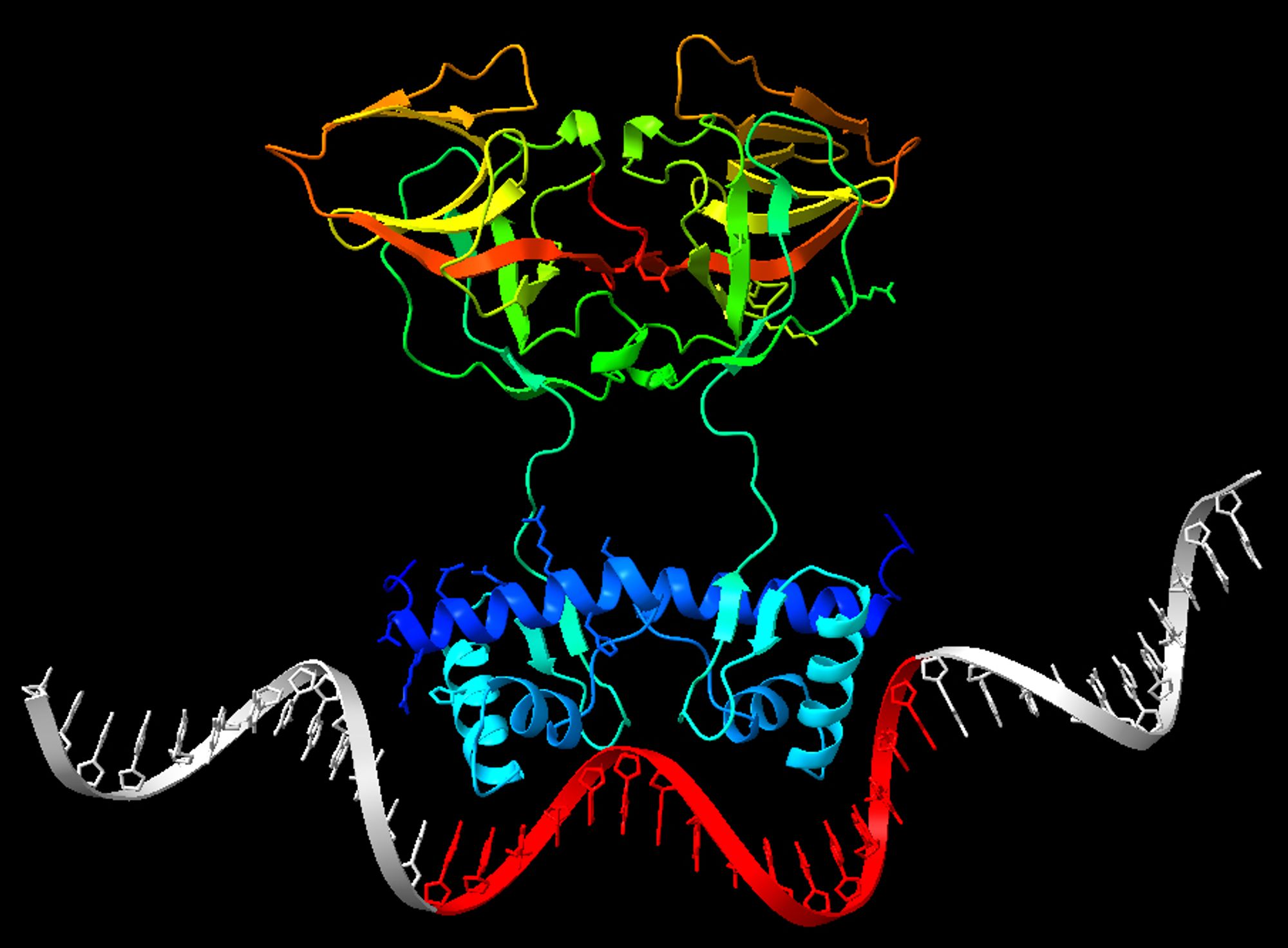
In 8/8 cases, the TF complex is perfect, and targets DNA using correctly the DNA binding domains. In 4/8 cases, the binding occurs precisely on my "randomized" LexA sites. Uppercase: from PWM; lowercase: random; underlined red: binding expected; highlighted yellow: bound. 6/

assembled correctly; (2) the LexA dimer contacts DNA with the known DNA binding domains in the DNA grooves; (3) it binds exactly on the "randomized" LexA sites (a pattern of 16 bp within the 40 bp). Here are the results on 8 sequences (we can't run >10 jobs/day at the moment) 5/
at which the site starts is also random (Uniform) so that the sequence may not be in the center. I input the sequence of LexA, and set "Copies" to 2 because LexA acts as a dimer. I then try AF3 on the sequences I generated. I consider the test passed if: (1) the dimer is 4/
sites I mean that I generate the DNA sequences by picking the base at each position according to the probabilities in the position weight matrix. You can get a seq that was not even among the examples. Then I embed each site into random DNA, for a total of 40 bp. The position 3/
of all, from what I read from Supplementary 2.5.2, AF3 was trained on Jaspar, which doesn't contain motifs for bacterial transcription factors! Please let me know if I'm missing some way in which bacterial TF motifs may have been available to AF3. Secondly, by "randomized" 2/
So, these these phages are unconventional in many ways. I'm grateful to all coauthors for this exciting collaboration. A lot of open questions remain and if some of you have a better idea than we do about what's going on, take the lead! Link to the paper: www.nature.com/articles/s41... [9/9]
One more peculiarity. When we talk about satellites in bacteria they are "satellite nucleic acids", not "satellite viruses" (which are common in plants). However, correct me if I'm wrong but these guys are bona fide satellite viruses because they encode for their own capsid proteins. [8/n]