Reading through a thread on latest conda TOS issue Conda TOS changed last year, and they're blocking IPs from larger nonprofit organizations (such as universities) with more than 200 connections/downloads. Connections will remain blocked until targeted organizations pay up. 💻🧬🧪
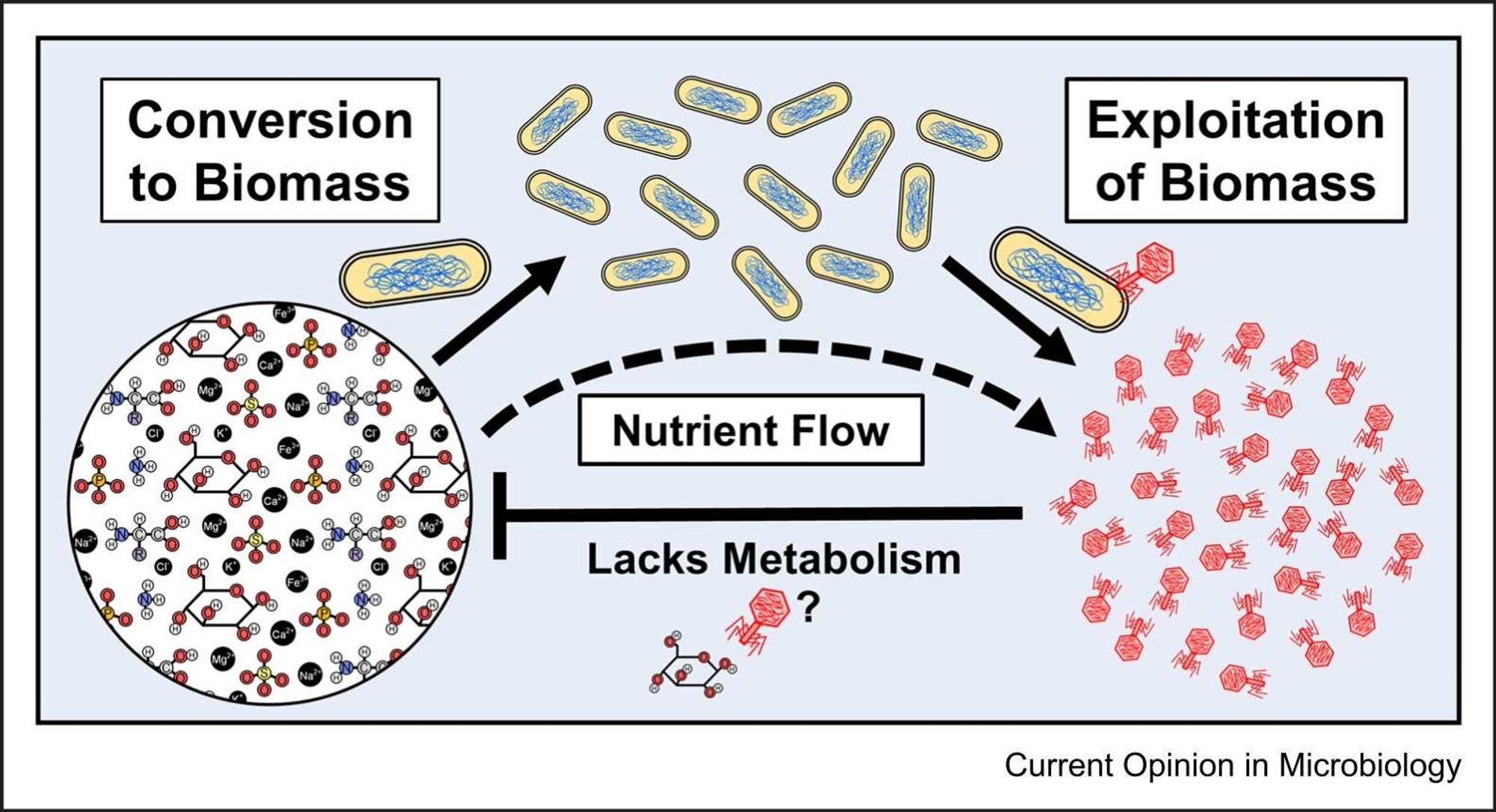
Two years ago we published about what we named "lytic deferment" in bacteriophages (doi.org/10.3389/fmic...doi.org/10.1016/j.mi... Food for thought. 🧬🦠🔬
Hey flagellum, shift into reverse gear! by Christoph — In a classical manual gearbox, a gearwheel is inserted between the countershaft and the drive shaft. This reverses the direction of rotation of the drive shaft. Flagella have found a smart... Read more > tinyurl.com/mrpeyfe8#MicroSky#CryoEM
![Screenshot from animation demonstrating gearing in the Salmonella flagellum. [Source: https://pubmed.ncbi.nlm.nih.gov/38459206/]](https://cdn.bsky.app/img/feed_fullsize/plain/did:plc:gudh3x7pgeh4uc6eee3zogtr/bafkreiemp4m2gpdh3whvhbkrow4olwurbba3ncymqhdetfdulmh5db4a3a@jpeg)
for their binding sites. Anyway, this is not a serious test. We'll need the code to go large-scale. It's just what I came up with given the limit of 10 jobs/day. After this limited experience with the webserver, I'm impressed! 🤯 END
also the others we have at least one perfect solution in 6/8 cases. I also tried with totally random DNA. As I expected it binds DNA the normal way (helices in groove) even if there's no LexA binding site. Not necessarily a "wrong" prediction given how bacterial TFs "search" 8/
Case 3 is particularly convincing because the site came out quite different from the consensus. It's not the classical CTGT-n8-ACAG, but AF3 predicts that the dimer binds there. AF3 can propose more than one model, but I was only looking at the first proposed. If I consider 7/

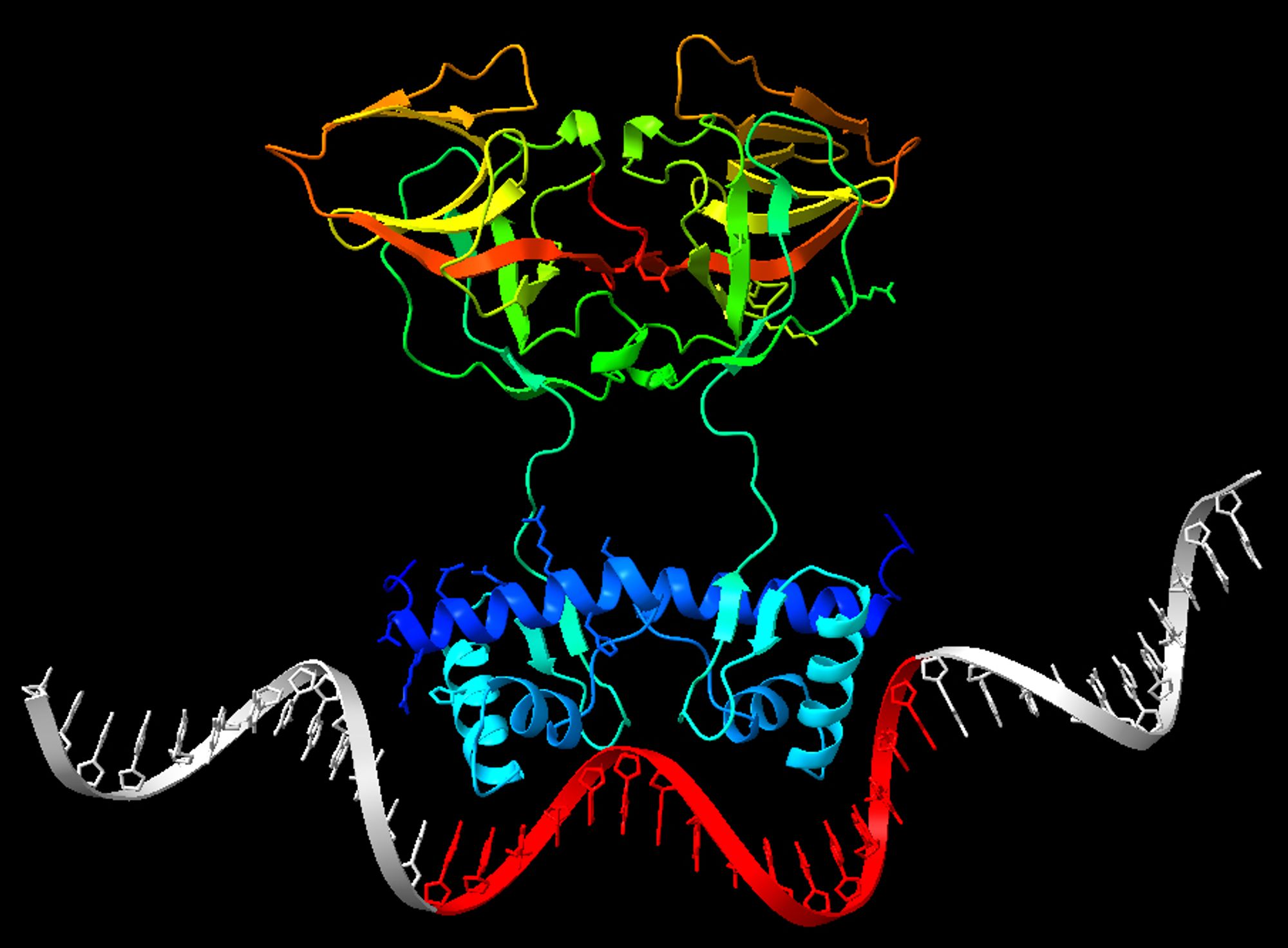
In 8/8 cases, the TF complex is perfect, and targets DNA using correctly the DNA binding domains. In 4/8 cases, the binding occurs precisely on my "randomized" LexA sites. Uppercase: from PWM; lowercase: random; underlined red: binding expected; highlighted yellow: bound. 6/

assembled correctly; (2) the LexA dimer contacts DNA with the known DNA binding domains in the DNA grooves; (3) it binds exactly on the "randomized" LexA sites (a pattern of 16 bp within the 40 bp). Here are the results on 8 sequences (we can't run >10 jobs/day at the moment) 5/
at which the site starts is also random (Uniform) so that the sequence may not be in the center. I input the sequence of LexA, and set "Copies" to 2 because LexA acts as a dimer. I then try AF3 on the sequences I generated. I consider the test passed if: (1) the dimer is 4/